Fluorine in PDB 7z2h: Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine
Enzymatic activity of Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine
All present enzymatic activity of Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine:
2.7.7.49; 2.7.7.7; 3.1.13.2; 3.1.26.13;
2.7.7.49; 2.7.7.7; 3.1.13.2; 3.1.26.13;
Other elements in 7z2h:
The structure of Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine also contains other interesting chemical elements:
| Chlorine | (Cl) | 1 atom |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine
(pdb code 7z2h). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine, PDB code: 7z2h:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine, PDB code: 7z2h:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 7z2h
Go back to
Fluorine binding site 1 out
of 3 in the Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine
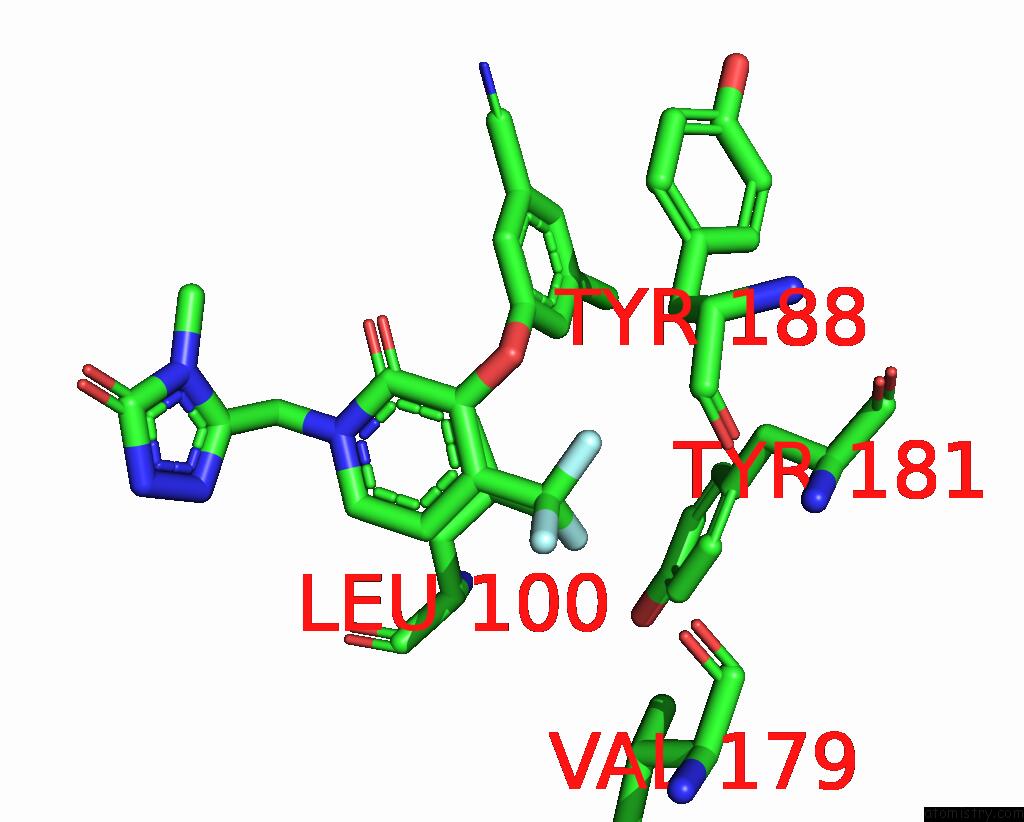
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine within 5.0Å range:
|
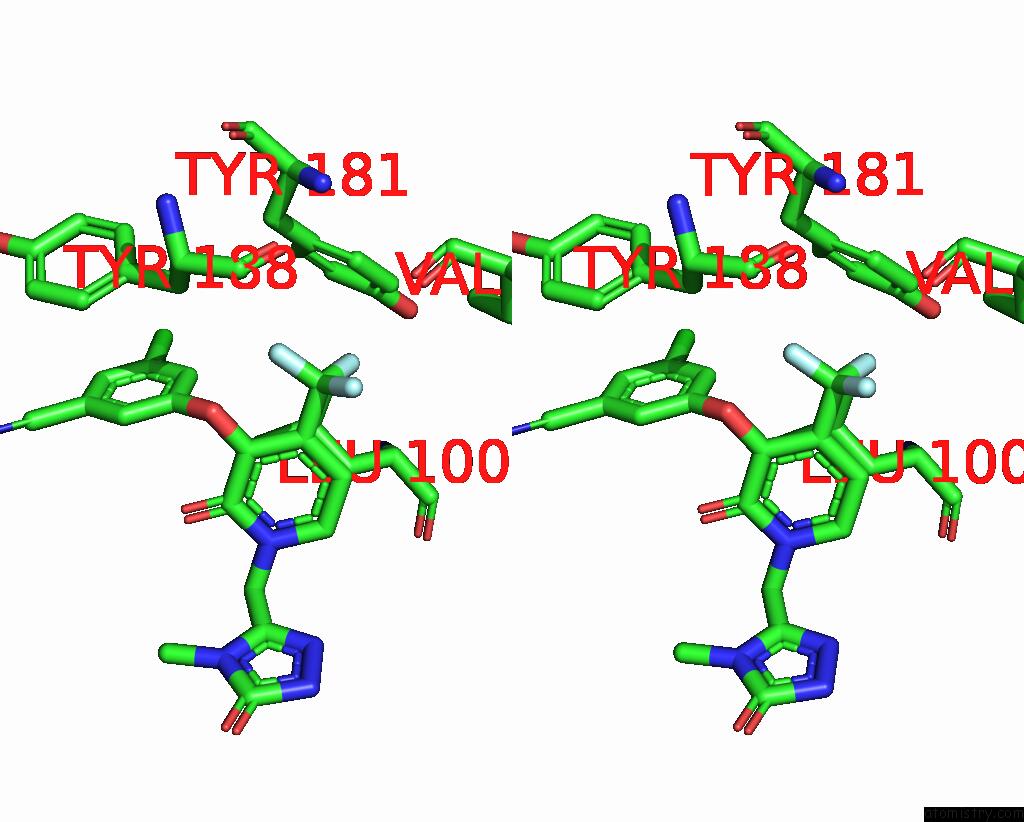
Fluorine binding site 2 out of 3 in 7z2h
Go back to
Fluorine binding site 2 out
of 3 in the Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine within 5.0Å range:
|
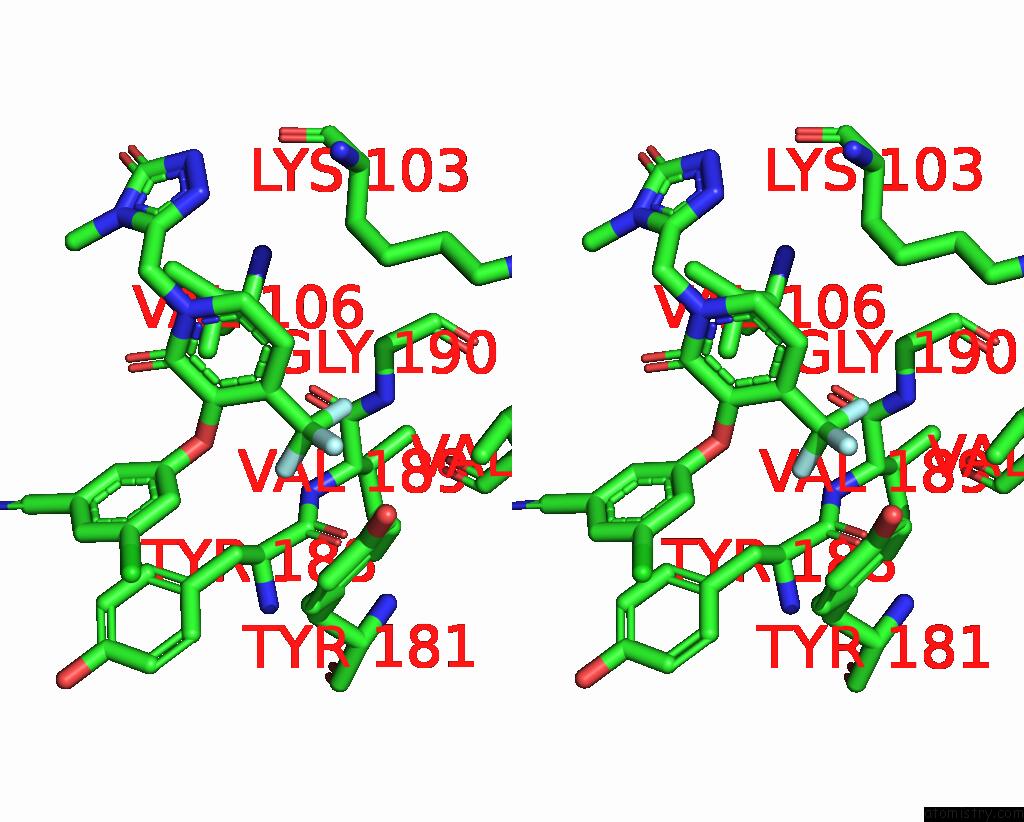
Fluorine binding site 3 out of 3 in 7z2h
Go back to
Fluorine binding site 3 out
of 3 in the Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine

Mono view
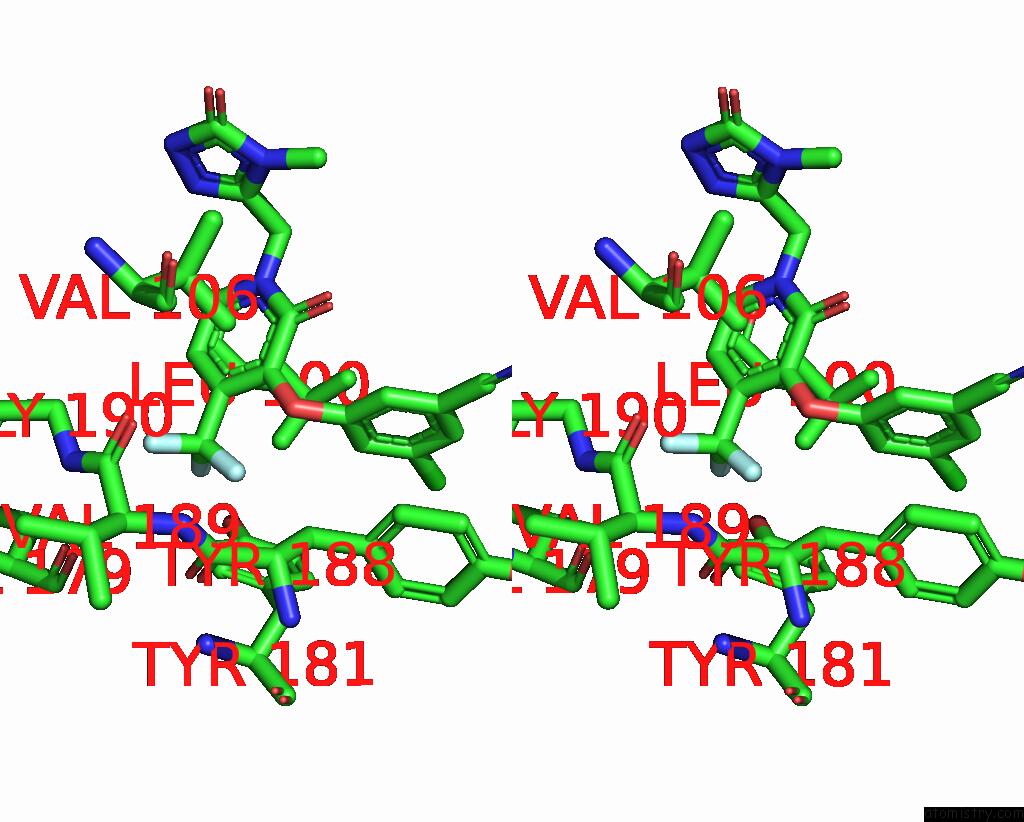
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Cryo-Em Structure of Nnrti Resistant M184I/E138K Mutant Hiv-1 Reverse Transcriptase with A Dna Aptamer in Complex with Doravirine within 5.0Å range:
|
Reference:
A.K.Singh,
B.De Wijngaert,
M.Bijnens,
K.Uyttersprot,
H.Nguyen,
S.E.Martinez,
D.Schols,
P.Herdewijn,
C.Pannecouque,
E.Arnold,
K.Das.
Cryo-Em Structures of Wild-Type and E138K/M184I Mutant Hiv-1 Rt/Dna Complexed with Inhibitors Doravirine and Rilpivirine. Proc.Natl.Acad.Sci.Usa V. 119 60119 2022.
ISSN: ESSN 1091-6490
PubMed: 35858448
DOI: 10.1073/PNAS.2203660119
Page generated: Fri Aug 2 16:03:47 2024
ISSN: ESSN 1091-6490
PubMed: 35858448
DOI: 10.1073/PNAS.2203660119
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF