Fluorine »
PDB 1xz3-1zzr »
1ysg »
Fluorine in PDB 1ysg: Solution Structure of the Anti-Apoptotic Protein Bcl-Xl in Complex with "Sar By uc(Nmr)" Ligands
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Solution Structure of the Anti-Apoptotic Protein Bcl-Xl in Complex with "Sar By uc(Nmr)" Ligands
(pdb code 1ysg). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total only one binding site of Fluorine was determined in the Solution Structure of the Anti-Apoptotic Protein Bcl-Xl in Complex with "Sar By uc(Nmr)" Ligands, PDB code: 1ysg:
In total only one binding site of Fluorine was determined in the Solution Structure of the Anti-Apoptotic Protein Bcl-Xl in Complex with "Sar By uc(Nmr)" Ligands, PDB code: 1ysg:
Fluorine binding site 1 out of 1 in 1ysg
Go back to
Fluorine binding site 1 out
of 1 in the Solution Structure of the Anti-Apoptotic Protein Bcl-Xl in Complex with "Sar By uc(Nmr)" Ligands
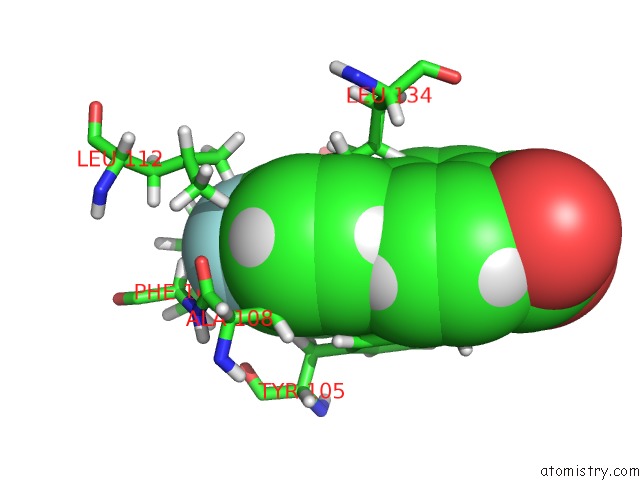
Mono view
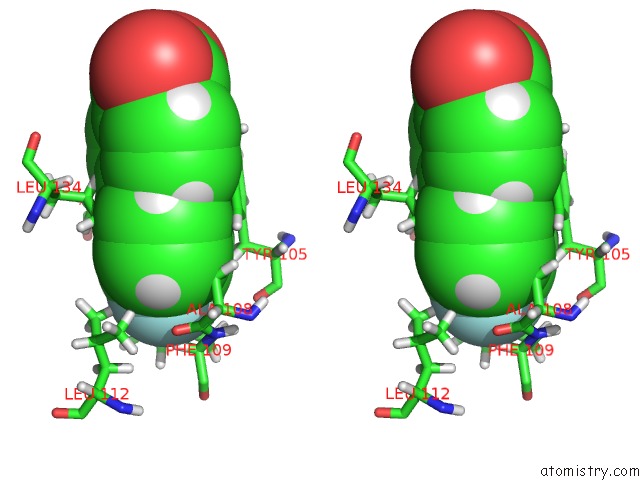
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Solution Structure of the Anti-Apoptotic Protein Bcl-Xl in Complex with "Sar By uc(Nmr)" Ligands within 5.0Å range:
|
Reference:
T.Oltersdorf,
S.W.Elmore,
A.R.Shoemaker,
R.C.Armstrong,
D.J.Augeri,
B.A.Belli,
M.Bruncko,
T.L.Deckwerth,
J.Dinges,
P.J.Hajduk,
M.K.Joseph,
S.Kitada,
S.J.Korsmeyer,
A.R.Kunzer,
A.Letai,
C.Li,
M.J.Mitten,
D.G.Nettesheim,
S.Ng,
P.M.Nimmer,
J.M.O'connor,
A.Oleksijew,
A.M.Petros,
J.C.Reed,
W.Shen,
S.K.Tahir,
C.B.Thompson,
K.J.Tomaselli,
B.Wang,
M.D.Wendt,
H.Zhang,
S.W.Fesik,
S.H.Rosenberg.
An Inhibitor of Bcl-2 Family Proteins Induces Regression of Solid Tumours Nature V. 435 677 2005.
ISSN: ISSN 0028-0836
PubMed: 15902208
DOI: 10.1038/NATURE03579
Page generated: Mon Jul 14 12:27:57 2025
ISSN: ISSN 0028-0836
PubMed: 15902208
DOI: 10.1038/NATURE03579
Last articles
Mg in 4GE0Mg in 4GBF
Mg in 4GDZ
Mg in 4GBU
Mg in 4GCY
Mg in 4GAD
Mg in 4GA3
Mg in 4G7O
Mg in 4G9B
Mg in 4G9O