Fluorine »
PDB 2q9p-2rbe »
2qhy »
Fluorine in PDB 2qhy: Crystal Structure of Protease Inhibitor, Mit-1-AC86 in Complex with Wild Type Hiv-1 Protease
Protein crystallography data
The structure of Crystal Structure of Protease Inhibitor, Mit-1-AC86 in Complex with Wild Type Hiv-1 Protease, PDB code: 2qhy
was solved by
C.A.Schiffer,
M.N.L.Nalam,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.18 / 1.85 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.670, 57.981, 61.541, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.9 / 19.7 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of Protease Inhibitor, Mit-1-AC86 in Complex with Wild Type Hiv-1 Protease
(pdb code 2qhy). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Crystal Structure of Protease Inhibitor, Mit-1-AC86 in Complex with Wild Type Hiv-1 Protease, PDB code: 2qhy:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Crystal Structure of Protease Inhibitor, Mit-1-AC86 in Complex with Wild Type Hiv-1 Protease, PDB code: 2qhy:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 2qhy
Go back to
Fluorine binding site 1 out
of 3 in the Crystal Structure of Protease Inhibitor, Mit-1-AC86 in Complex with Wild Type Hiv-1 Protease
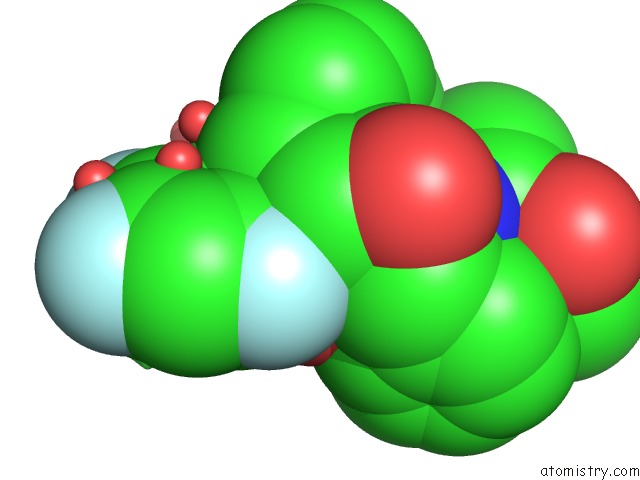
Mono view
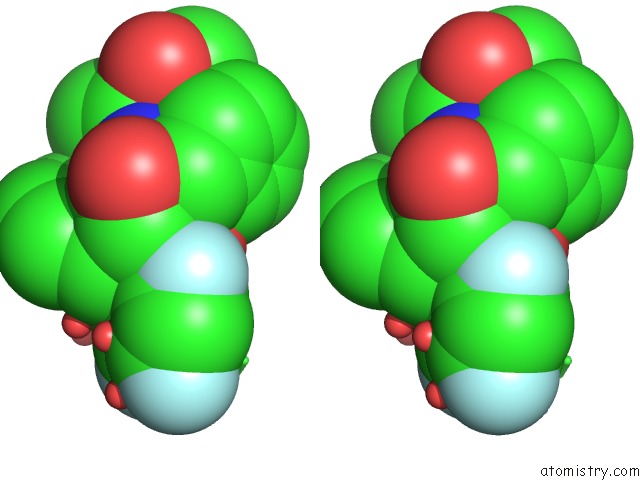
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of Protease Inhibitor, Mit-1-AC86 in Complex with Wild Type Hiv-1 Protease within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 2qhy
Go back to
Fluorine binding site 2 out
of 3 in the Crystal Structure of Protease Inhibitor, Mit-1-AC86 in Complex with Wild Type Hiv-1 Protease
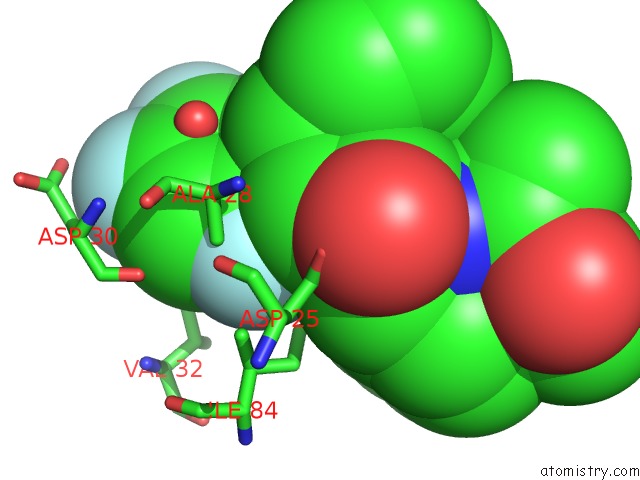
Mono view
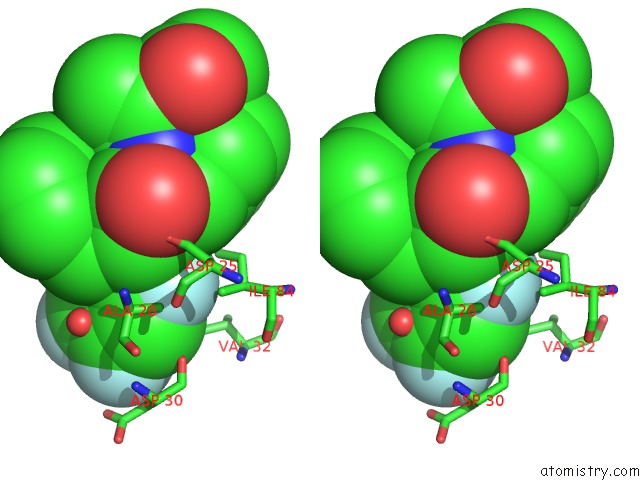
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of Protease Inhibitor, Mit-1-AC86 in Complex with Wild Type Hiv-1 Protease within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 2qhy
Go back to
Fluorine binding site 3 out
of 3 in the Crystal Structure of Protease Inhibitor, Mit-1-AC86 in Complex with Wild Type Hiv-1 Protease
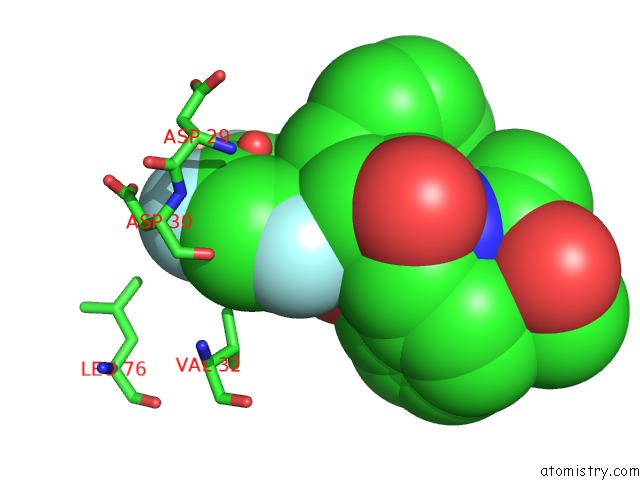
Mono view
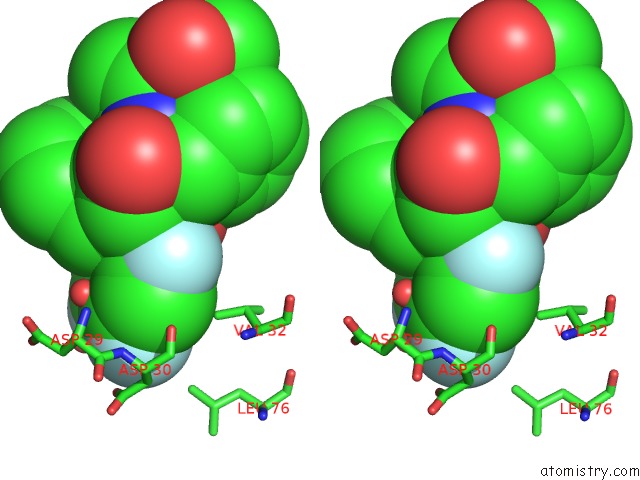
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Crystal Structure of Protease Inhibitor, Mit-1-AC86 in Complex with Wild Type Hiv-1 Protease within 5.0Å range:
|
Reference:
M.D.Altman,
A.Ali,
G.S.Reddy,
M.N.Nalam,
S.G.Anjum,
H.Cao,
S.Chellappan,
V.Kairys,
M.X.Fernandes,
M.K.Gilson,
C.A.Schiffer,
T.M.Rana,
B.Tidor.
Hiv-1 Protease Inhibitors From Inverse Design in the Substrate Envelope Exhibit Subnanomolar Binding to Drug-Resistant Variants. J.Am.Chem.Soc. V. 130 6099 2008.
ISSN: ISSN 0002-7863
PubMed: 18412349
DOI: 10.1021/JA076558P
Page generated: Mon Jul 14 14:09:55 2025
ISSN: ISSN 0002-7863
PubMed: 18412349
DOI: 10.1021/JA076558P
Last articles
K in 9HKXK in 9HKY
K in 9HKW
K in 9HFO
K in 9HFN
K in 9HI3
K in 9HFM
K in 9HAC
K in 9GXH
K in 9HAG