Fluorine »
PDB 4xyf-4ymo »
4y34 »
Fluorine in PDB 4y34: Crystal Structure of Coxsackievirus B3 3D Polymerase in Complex with Gpc-N143
Protein crystallography data
The structure of Crystal Structure of Coxsackievirus B3 3D Polymerase in Complex with Gpc-N143, PDB code: 4y34
was solved by
L.Vives-Adrian,
C.Ferrer-Orta,
N.Cerdaguer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 72.20 / 2.70 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.620, 74.620, 288.430, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.4 / 24.1 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of Coxsackievirus B3 3D Polymerase in Complex with Gpc-N143
(pdb code 4y34). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total only one binding site of Fluorine was determined in the Crystal Structure of Coxsackievirus B3 3D Polymerase in Complex with Gpc-N143, PDB code: 4y34:
In total only one binding site of Fluorine was determined in the Crystal Structure of Coxsackievirus B3 3D Polymerase in Complex with Gpc-N143, PDB code: 4y34:
Fluorine binding site 1 out of 1 in 4y34
Go back to
Fluorine binding site 1 out
of 1 in the Crystal Structure of Coxsackievirus B3 3D Polymerase in Complex with Gpc-N143
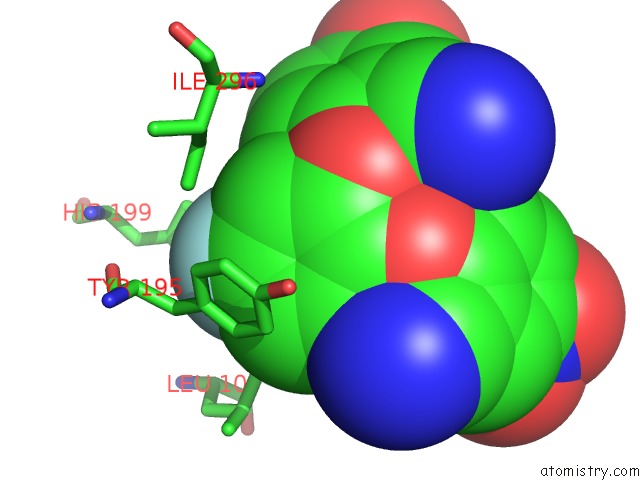
Mono view
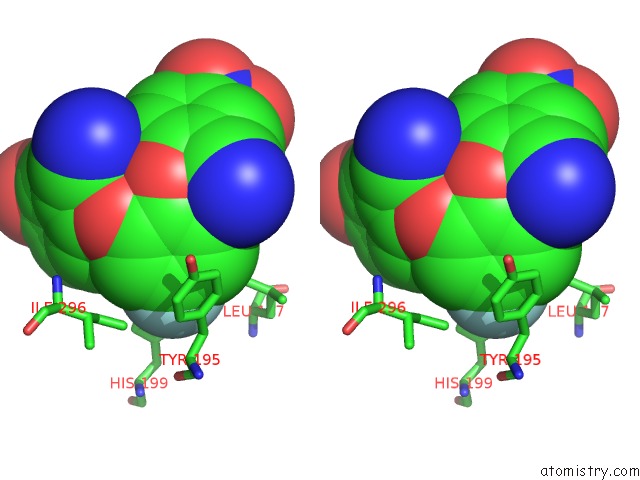
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of Coxsackievirus B3 3D Polymerase in Complex with Gpc-N143 within 5.0Å range:
|
Reference:
L.Van Der Linden,
L.Vives-Adrian,
B.Selisko,
C.Ferrer-Orta,
X.Liu,
K.Lanke,
R.Ulferts,
A.M.De Palma,
F.Tanchis,
N.Goris,
D.Lefevbre,
K.De Clercq,
P.Leyssen,
C.Lacroix,
G.Purstinger,
B.Coutard,
B.Canard,
D.D.Boehr,
J.J.Arnold,
C.E.Cameron,
N.Verdaguer,
J.Neyts,
F.Van Kuppeveld.
The Rna Template Channel of the Rna-Dependent Rna Polymerase As A Target For Development of Antiviral Therapy of Multiple Genera Within A Virus Family To Be Published.
Page generated: Tue Jul 15 01:37:50 2025
Last articles
Mg in 5SHQMg in 5SHP
Mg in 5SHN
Mg in 5SHO
Mg in 5SHL
Mg in 5SHM
Mg in 5SHK
Mg in 5SHJ
Mg in 5SHI
Mg in 5SHH