Fluorine »
PDB 5tw3-5ug8 »
5uey »
Fluorine in PDB 5uey: BRD4_BD2_A-1412838
Protein crystallography data
The structure of BRD4_BD2_A-1412838, PDB code: 5uey
was solved by
C.H.Park,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.38 / 2.41 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 54.185, 74.627, 33.376, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 27 / 31 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the BRD4_BD2_A-1412838
(pdb code 5uey). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the BRD4_BD2_A-1412838, PDB code: 5uey:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the BRD4_BD2_A-1412838, PDB code: 5uey:
Jump to Fluorine binding site number: 1; 2;
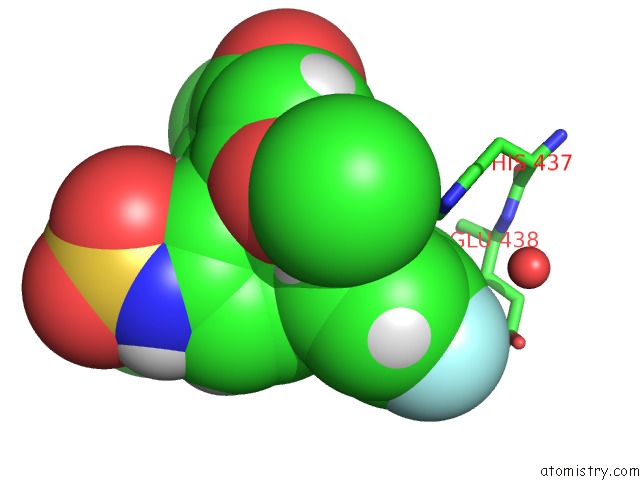
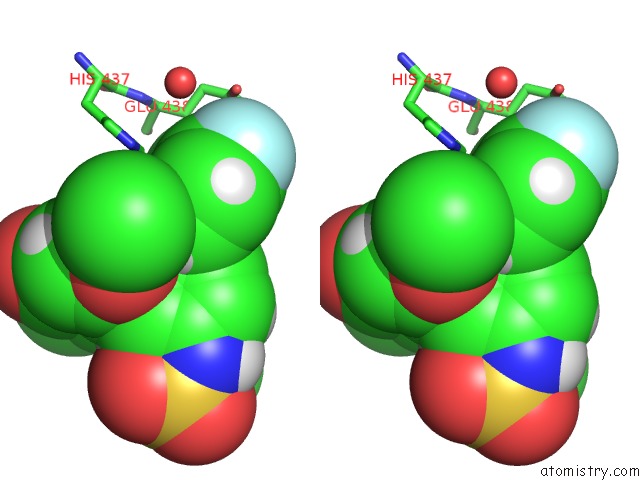
Fluorine binding site 1 out of 2 in 5uey
Go back to
Fluorine binding site 1 out
of 2 in the BRD4_BD2_A-1412838
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of BRD4_BD2_A-1412838 within 5.0Å range:
|
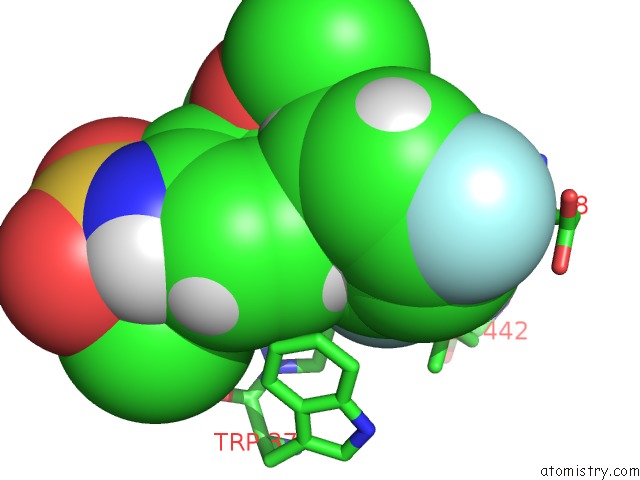
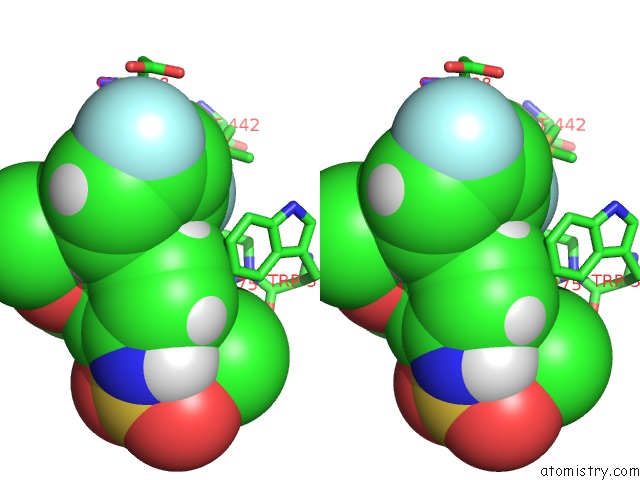
Fluorine binding site 2 out of 2 in 5uey
Go back to
Fluorine binding site 2 out
of 2 in the BRD4_BD2_A-1412838
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of BRD4_BD2_A-1412838 within 5.0Å range:
|
Reference:
L.Wang,
J.K.Pratt,
T.Soltwedel,
G.S.Sheppard,
S.D.Fidanze,
D.Liu,
L.A.Hasvold,
R.A.Mantei,
J.H.Holms,
W.J.Mcclellan,
M.D.Wendt,
C.Wada,
R.Frey,
T.M.Hansen,
R.Hubbard,
C.H.Park,
L.Li,
T.J.Magoc,
D.H.Albert,
X.Lin,
S.E.Warder,
P.Kovar,
X.Huang,
D.Wilcox,
R.Wang,
G.Rajaraman,
A.M.Petros,
C.W.Hutchins,
S.C.Panchal,
C.Sun,
S.W.Elmore,
Y.Shen,
W.M.Kati,
K.F.Mcdaniel.
Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles As Potent Bromodomain and Extra-Terminal Domain (Bet) Family Bromodomain Inhibitors. J. Med. Chem. V. 60 3828 2017.
ISSN: ISSN 1520-4804
PubMed: 28368119
DOI: 10.1021/ACS.JMEDCHEM.7B00017
Page generated: Tue Jul 15 08:12:54 2025
ISSN: ISSN 1520-4804
PubMed: 28368119
DOI: 10.1021/ACS.JMEDCHEM.7B00017
Last articles
Fe in 8XLSFe in 8XLP
Fe in 8XMQ
Fe in 8XMP
Fe in 8XIW
Fe in 8XCM
Fe in 8XCN
Fe in 8XHT
Fe in 8XHP
Fe in 8XFI