Fluorine »
PDB 6b3e-6bkw »
6b7e »
Fluorine in PDB 6b7e: Crystal Structure of E.Coli Phosphopantetheine Adenylyltransferase (Ppat/Coad) in Complex with (R)-4-(5-(Difluoromethyl)-1H-Imidazol-1- Yl)-3,3-Dimethylisochroman-1-One
Enzymatic activity of Crystal Structure of E.Coli Phosphopantetheine Adenylyltransferase (Ppat/Coad) in Complex with (R)-4-(5-(Difluoromethyl)-1H-Imidazol-1- Yl)-3,3-Dimethylisochroman-1-One
All present enzymatic activity of Crystal Structure of E.Coli Phosphopantetheine Adenylyltransferase (Ppat/Coad) in Complex with (R)-4-(5-(Difluoromethyl)-1H-Imidazol-1- Yl)-3,3-Dimethylisochroman-1-One:
2.7.7.3;
2.7.7.3;
Protein crystallography data
The structure of Crystal Structure of E.Coli Phosphopantetheine Adenylyltransferase (Ppat/Coad) in Complex with (R)-4-(5-(Difluoromethyl)-1H-Imidazol-1- Yl)-3,3-Dimethylisochroman-1-One, PDB code: 6b7e
was solved by
A.W.Proudfoot,
D.Bussiere,
A.Lingel,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.29 / 2.10 |
| Space group | I 2 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 135.480, 135.480, 135.480, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.1 / 20.6 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of E.Coli Phosphopantetheine Adenylyltransferase (Ppat/Coad) in Complex with (R)-4-(5-(Difluoromethyl)-1H-Imidazol-1- Yl)-3,3-Dimethylisochroman-1-One
(pdb code 6b7e). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Crystal Structure of E.Coli Phosphopantetheine Adenylyltransferase (Ppat/Coad) in Complex with (R)-4-(5-(Difluoromethyl)-1H-Imidazol-1- Yl)-3,3-Dimethylisochroman-1-One, PDB code: 6b7e:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Crystal Structure of E.Coli Phosphopantetheine Adenylyltransferase (Ppat/Coad) in Complex with (R)-4-(5-(Difluoromethyl)-1H-Imidazol-1- Yl)-3,3-Dimethylisochroman-1-One, PDB code: 6b7e:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 6b7e
Go back to
Fluorine binding site 1 out
of 2 in the Crystal Structure of E.Coli Phosphopantetheine Adenylyltransferase (Ppat/Coad) in Complex with (R)-4-(5-(Difluoromethyl)-1H-Imidazol-1- Yl)-3,3-Dimethylisochroman-1-One
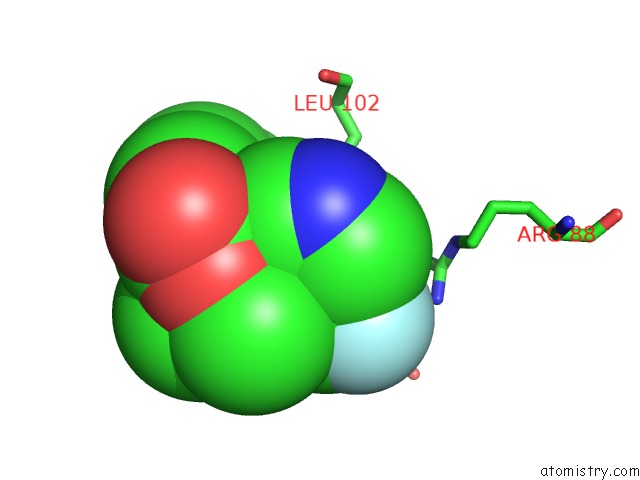
Mono view
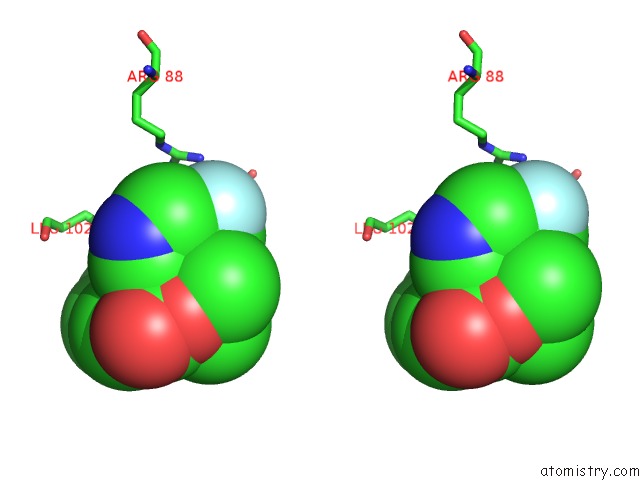
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of E.Coli Phosphopantetheine Adenylyltransferase (Ppat/Coad) in Complex with (R)-4-(5-(Difluoromethyl)-1H-Imidazol-1- Yl)-3,3-Dimethylisochroman-1-One within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 6b7e
Go back to
Fluorine binding site 2 out
of 2 in the Crystal Structure of E.Coli Phosphopantetheine Adenylyltransferase (Ppat/Coad) in Complex with (R)-4-(5-(Difluoromethyl)-1H-Imidazol-1- Yl)-3,3-Dimethylisochroman-1-One
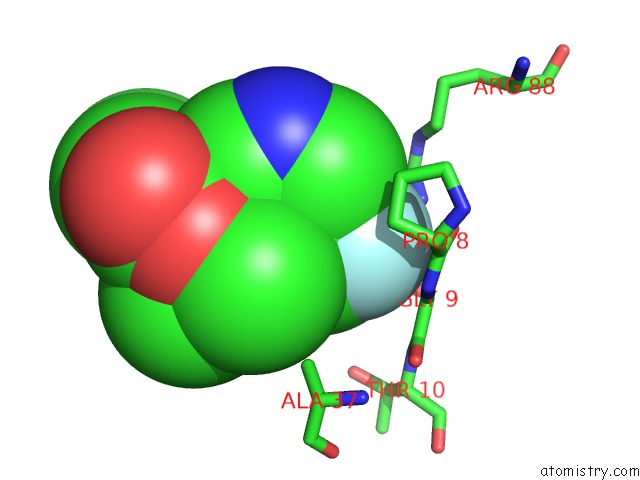
Mono view
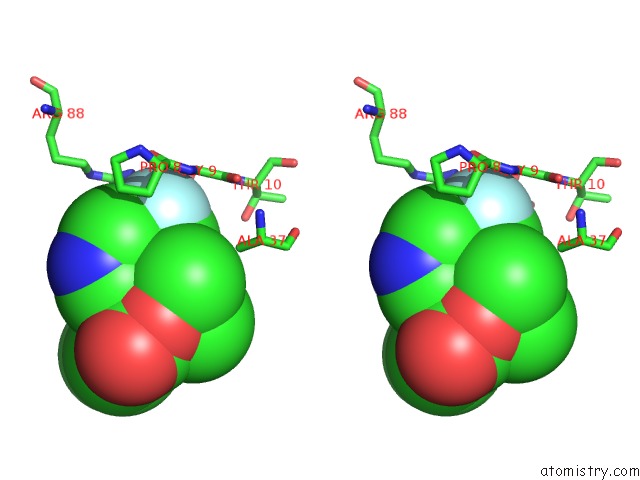
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of E.Coli Phosphopantetheine Adenylyltransferase (Ppat/Coad) in Complex with (R)-4-(5-(Difluoromethyl)-1H-Imidazol-1- Yl)-3,3-Dimethylisochroman-1-One within 5.0Å range:
|
Reference:
A.Proudfoot,
D.E.Bussiere,
A.Lingel.
High-Confidence Protein-Ligand Complex Modeling By uc(Nmr)-Guided Docking Enables Early Hit Optimization. J. Am. Chem. Soc. V. 139 17824 2017.
ISSN: ESSN 1520-5126
PubMed: 29190085
DOI: 10.1021/JACS.7B07171
Page generated: Tue Jul 15 09:58:12 2025
ISSN: ESSN 1520-5126
PubMed: 29190085
DOI: 10.1021/JACS.7B07171
Last articles
Mg in 3B04Mg in 3B03
Mg in 3AYZ
Mg in 3AYX
Mg in 3AY9
Mg in 3AXM
Mg in 3AWD
Mg in 3AXK
Mg in 3AUO
Mg in 3AV3