Fluorine »
PDB 6ffb-6g0l »
6fz4 »
Fluorine in PDB 6fz4: Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution
Protein crystallography data
The structure of Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution, PDB code: 6fz4
was solved by
J.S.Kastrup,
K.Frydenvang,
S.Mollerud,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.38 / 1.85 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 88.867, 88.867, 157.145, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.7 / 21.6 |
Other elements in 6fz4:
The structure of Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution also contains other interesting chemical elements:
| Chlorine | (Cl) | 1 atom |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution
(pdb code 6fz4). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 4 binding sites of Fluorine where determined in the Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution, PDB code: 6fz4:
Jump to Fluorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Fluorine where determined in the Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution, PDB code: 6fz4:
Jump to Fluorine binding site number: 1; 2; 3; 4;
Fluorine binding site 1 out of 4 in 6fz4
Go back to
Fluorine binding site 1 out
of 4 in the Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution
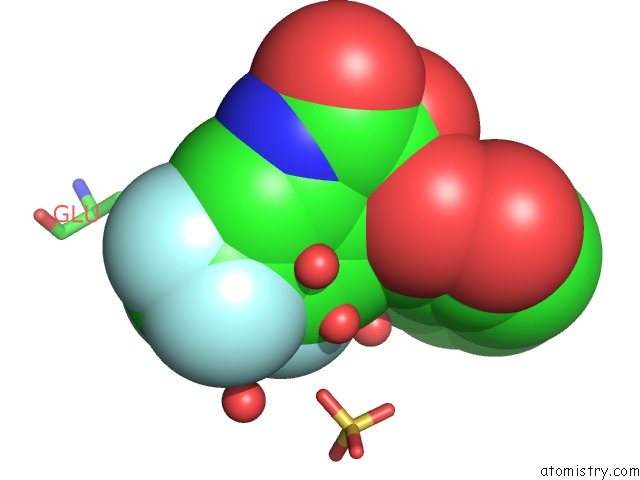
Mono view
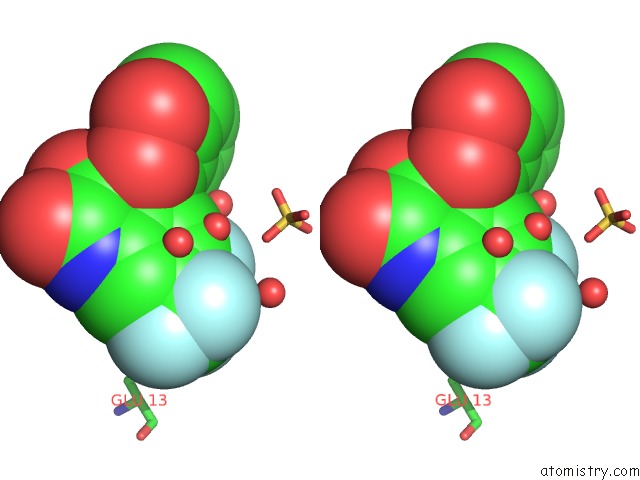
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution within 5.0Å range:
|
Fluorine binding site 2 out of 4 in 6fz4
Go back to
Fluorine binding site 2 out
of 4 in the Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution

Mono view
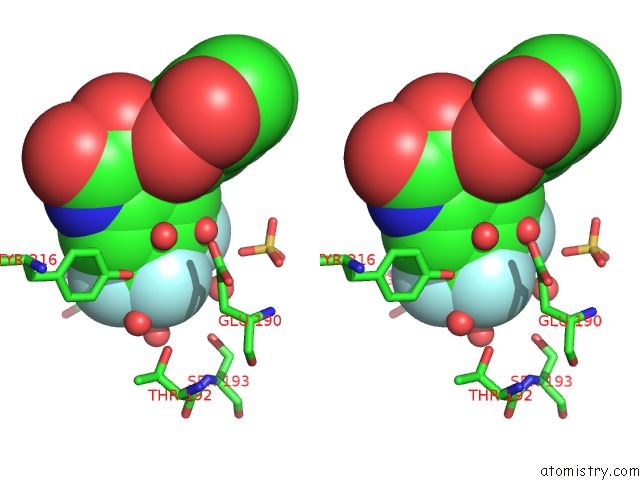
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution within 5.0Å range:
|
Fluorine binding site 3 out of 4 in 6fz4
Go back to
Fluorine binding site 3 out
of 4 in the Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution within 5.0Å range:
|
Fluorine binding site 4 out of 4 in 6fz4
Go back to
Fluorine binding site 4 out
of 4 in the Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 4 of Structure of GLUK1 Ligand-Binding Domain in Complex with N-(7-Fluoro- 2,3-Dioxo-6-(Trifluoromethyl)-3,4-Dihydroquinoxalin-1(2H)-Yl)-2- Hydroxybenzamide at 1.85 A Resolution within 5.0Å range:
|
Reference:
J.Pallesen,
S.Mollerud,
K.Frydenvang,
D.S.Pickering,
J.Bornholdt,
B.Nielsen,
D.Pasini,
L.Han,
L.Marconi,
J.S.Kastrup,
T.N.Johansen.
N1-Substituted Quinoxaline-2,3-Diones As Kainate Receptor Antagonists: X-Ray Crystallography, Structure-Affinity Relationships, and in Vitro Pharmacology. Acs Chem Neurosci V. 10 1841 2019.
ISSN: ESSN 1948-7193
PubMed: 30620174
DOI: 10.1021/ACSCHEMNEURO.8B00726
Page generated: Tue Jul 15 11:42:26 2025
ISSN: ESSN 1948-7193
PubMed: 30620174
DOI: 10.1021/ACSCHEMNEURO.8B00726
Last articles
Mg in 1FE1Mg in 1FNT
Mg in 1FNQ
Mg in 1FNP
Mg in 1FNN
Mg in 1FNM
Mg in 1FMW
Mg in 1FMS
Mg in 1FIU
Mg in 1FMQ