Fluorine »
PDB 6oaq-6onv »
6ogr »
Fluorine in PDB 6ogr: X-Ray Crystal Structure of Darunavir-Resistant Hiv-1 Protease (P30) in Complex with Grl-142
Protein crystallography data
The structure of X-Ray Crystal Structure of Darunavir-Resistant Hiv-1 Protease (P30) in Complex with Grl-142, PDB code: 6ogr
was solved by
H.Bulut,
S.I.Hattori,
H.Aoki-Ogata,
H.Hayashi,
M.Aoki,
A.K.Ghosh,
H.Mitsuya,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 54.42 / 1.28 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 62.845, 62.845, 81.808, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20.7 / 22.9 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the X-Ray Crystal Structure of Darunavir-Resistant Hiv-1 Protease (P30) in Complex with Grl-142
(pdb code 6ogr). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the X-Ray Crystal Structure of Darunavir-Resistant Hiv-1 Protease (P30) in Complex with Grl-142, PDB code: 6ogr:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the X-Ray Crystal Structure of Darunavir-Resistant Hiv-1 Protease (P30) in Complex with Grl-142, PDB code: 6ogr:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 6ogr
Go back to
Fluorine binding site 1 out
of 2 in the X-Ray Crystal Structure of Darunavir-Resistant Hiv-1 Protease (P30) in Complex with Grl-142
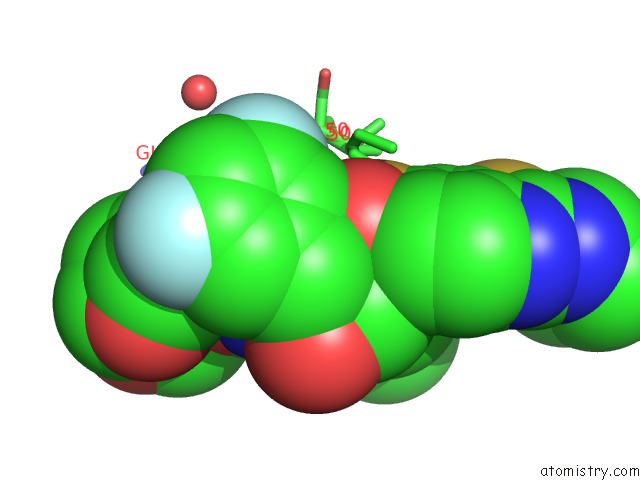
Mono view
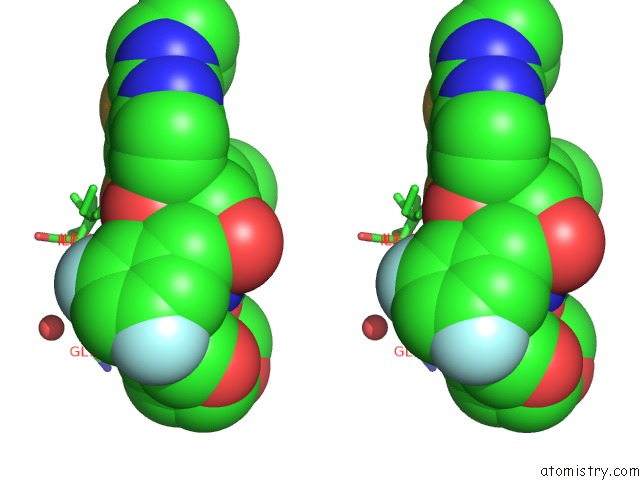
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of X-Ray Crystal Structure of Darunavir-Resistant Hiv-1 Protease (P30) in Complex with Grl-142 within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 6ogr
Go back to
Fluorine binding site 2 out
of 2 in the X-Ray Crystal Structure of Darunavir-Resistant Hiv-1 Protease (P30) in Complex with Grl-142
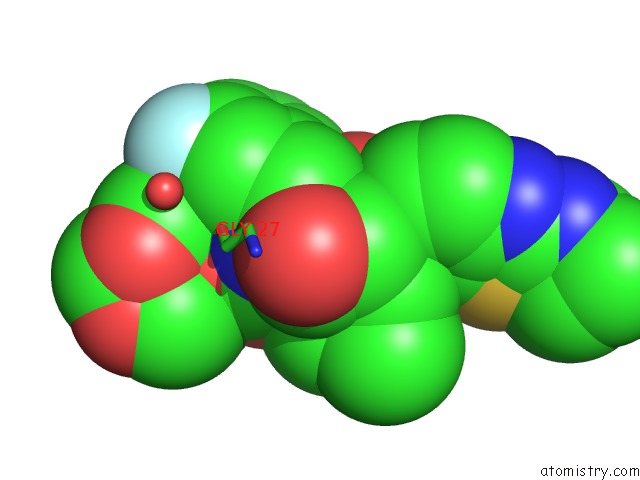
Mono view
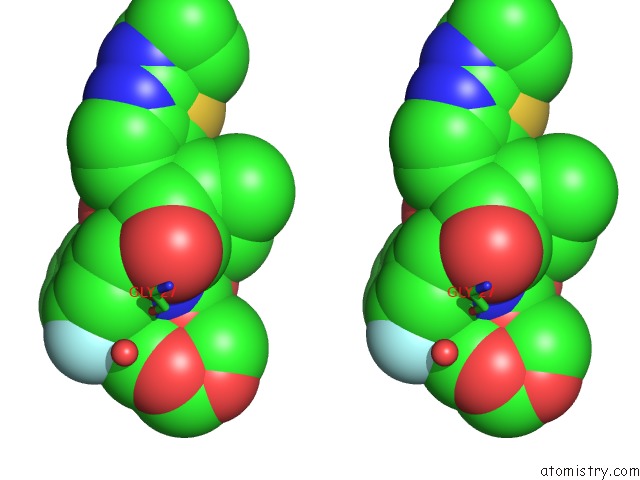
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of X-Ray Crystal Structure of Darunavir-Resistant Hiv-1 Protease (P30) in Complex with Grl-142 within 5.0Å range:
|
Reference:
H.Bulut,
S.I.Hattori,
H.Aoki-Ogata,
H.Hayashi,
M.Aoki,
A.K.Ghosh,
H.Mitsuya.
Novel Hiv-1 Protease Inhibitors, Grl-142 and Its Analogs, Markedly Adapt to the Structural Plasticity of Hiv-1 Protease and Exerts Extreme Potency with High Genetic Barrier To Be Published.
Page generated: Tue Jul 15 14:10:54 2025
Last articles
Na in 3DDKNa in 3DFH
Na in 3DEB
Na in 3DAV
Na in 3DDR
Na in 3DBO
Na in 3DC7
Na in 3D97
Na in 3DA9
Na in 3D9R