Fluorine »
PDB 6pfa-6q0t »
6pzo »
Fluorine in PDB 6pzo: Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153
Enzymatic activity of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153
All present enzymatic activity of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153:
3.5.1.98;
3.5.1.98;
Protein crystallography data
The structure of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153, PDB code: 6pzo
was solved by
J.D.Osko,
D.W.Christianson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 66.88 / 1.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.810, 92.580, 96.730, 90.00, 90.00, 90.00 |
| R / Rfree (%) | n/a / n/a |
Other elements in 6pzo:
The structure of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153 also contains other interesting chemical elements:
| Potassium | (K) | 4 atoms |
| Zinc | (Zn) | 2 atoms |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153
(pdb code 6pzo). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 6 binding sites of Fluorine where determined in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153, PDB code: 6pzo:
Jump to Fluorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Fluorine where determined in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153, PDB code: 6pzo:
Jump to Fluorine binding site number: 1; 2; 3; 4; 5; 6;
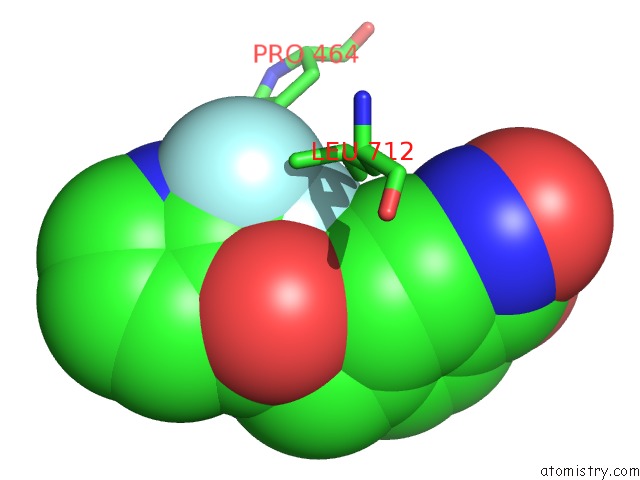
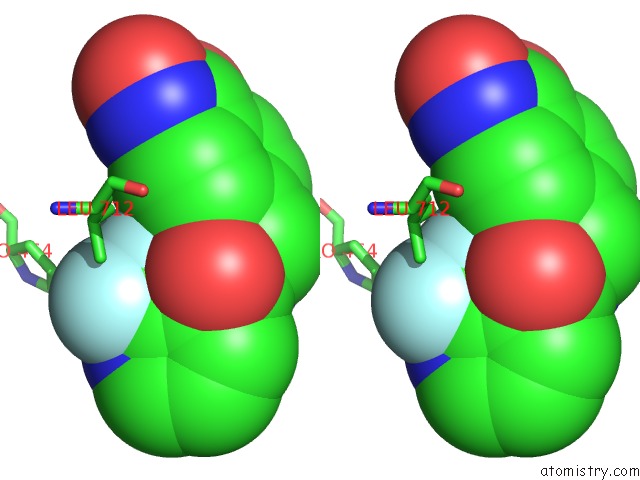
Fluorine binding site 1 out of 6 in 6pzo
Go back to
Fluorine binding site 1 out
of 6 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153 within 5.0Å range:
|
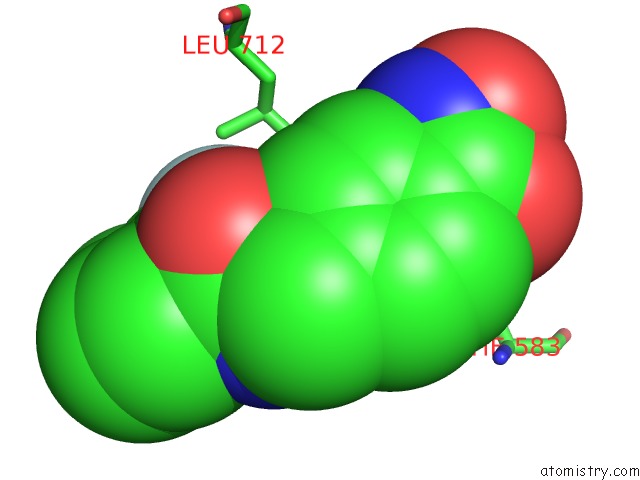
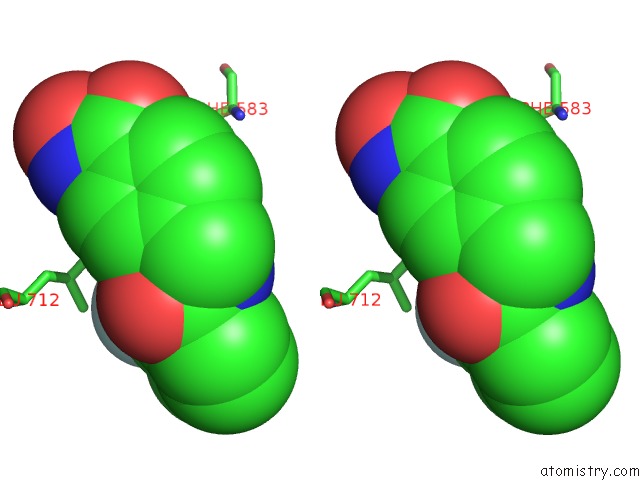
Fluorine binding site 2 out of 6 in 6pzo
Go back to
Fluorine binding site 2 out
of 6 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153 within 5.0Å range:
|
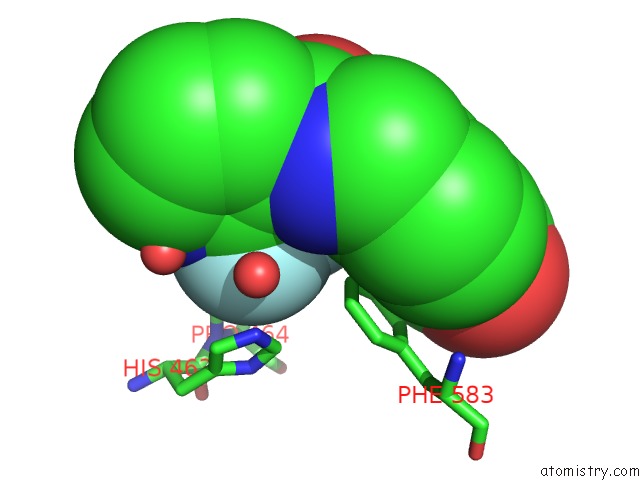
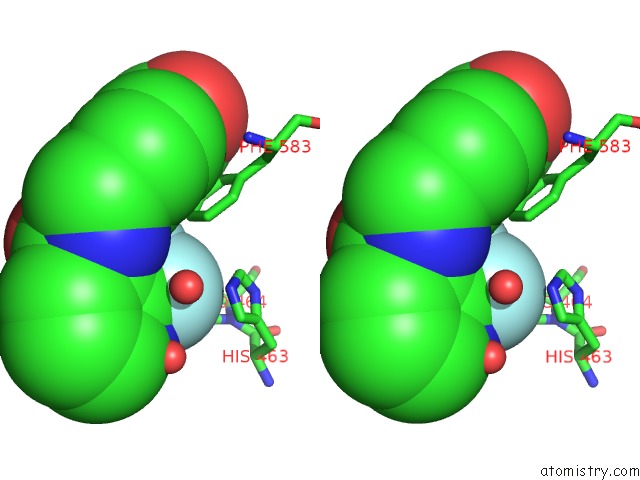
Fluorine binding site 3 out of 6 in 6pzo
Go back to
Fluorine binding site 3 out
of 6 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153 within 5.0Å range:
|
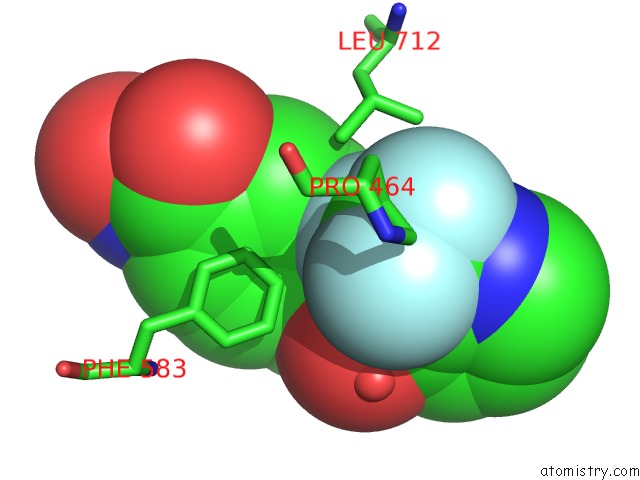
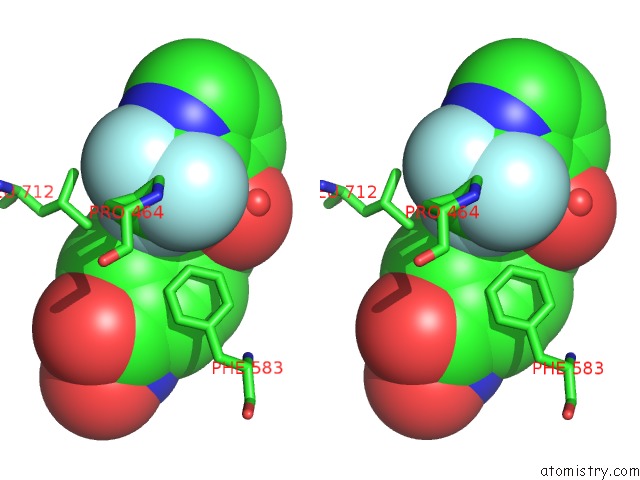
Fluorine binding site 4 out of 6 in 6pzo
Go back to
Fluorine binding site 4 out
of 6 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 4 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153 within 5.0Å range:
|
Fluorine binding site 5 out of 6 in 6pzo
Go back to
Fluorine binding site 5 out
of 6 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153
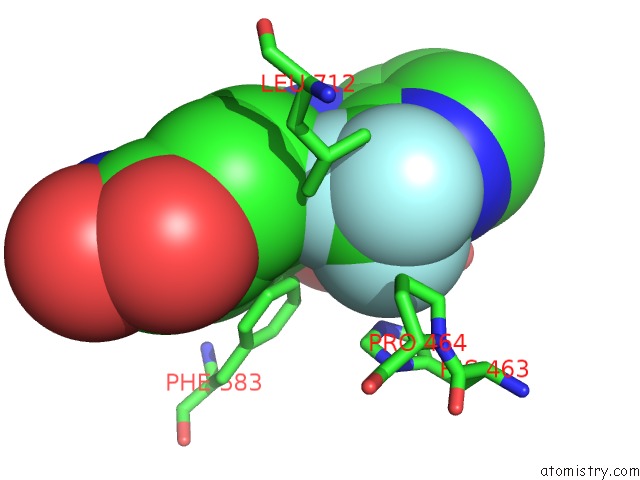
Mono view
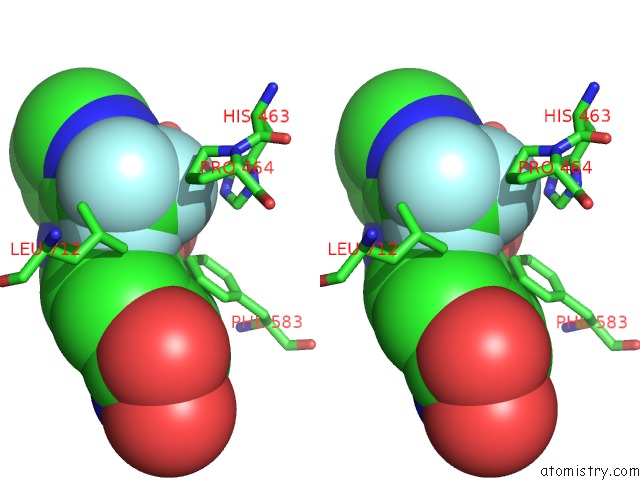
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 5 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153 within 5.0Å range:
|
Fluorine binding site 6 out of 6 in 6pzo
Go back to
Fluorine binding site 6 out
of 6 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153
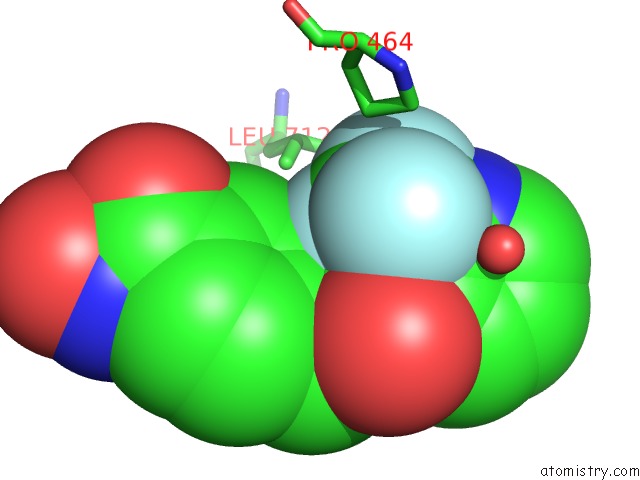
Mono view
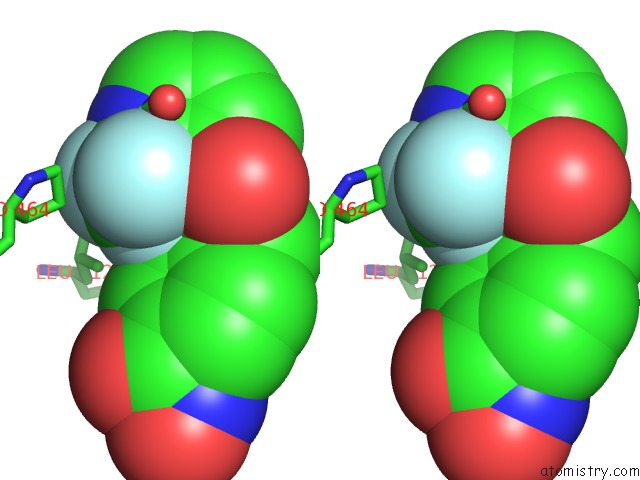
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 6 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed with Yx-153 within 5.0Å range:
|
Reference:
J.D.Osko,
N.J.Porter,
P.A.Narayana Reddy,
Y.C.Xiao,
J.Rokka,
M.Jung,
J.M.Hooker,
J.M.Salvino,
D.W.Christianson.
Exploring Structural Determinants of Inhibitor Affinity and Selectivity in Complexes with Histone Deacetylase 6. J.Med.Chem. V. 63 295 2020.
ISSN: ISSN 0022-2623
PubMed: 31793776
DOI: 10.1021/ACS.JMEDCHEM.9B01540
Page generated: Tue Jul 15 14:52:49 2025
ISSN: ISSN 0022-2623
PubMed: 31793776
DOI: 10.1021/ACS.JMEDCHEM.9B01540
Last articles
I in 4P4YI in 4P4W
I in 4OA5
I in 4OWA
I in 4P4V
I in 4OWC
I in 4P0D
I in 4OP3
I in 4OP2
I in 4OUC