Fluorine »
PDB 6rln-6rzh »
6roi »
Fluorine in PDB 6roi: Cryo-Em Structure of the Partially Activated DRS2P-CDC50P
Enzymatic activity of Cryo-Em Structure of the Partially Activated DRS2P-CDC50P
All present enzymatic activity of Cryo-Em Structure of the Partially Activated DRS2P-CDC50P:
7.6.2.1;
7.6.2.1;
Other elements in 6roi:
The structure of Cryo-Em Structure of the Partially Activated DRS2P-CDC50P also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Cryo-Em Structure of the Partially Activated DRS2P-CDC50P
(pdb code 6roi). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Cryo-Em Structure of the Partially Activated DRS2P-CDC50P, PDB code: 6roi:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Cryo-Em Structure of the Partially Activated DRS2P-CDC50P, PDB code: 6roi:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 6roi
Go back to
Fluorine binding site 1 out
of 3 in the Cryo-Em Structure of the Partially Activated DRS2P-CDC50P
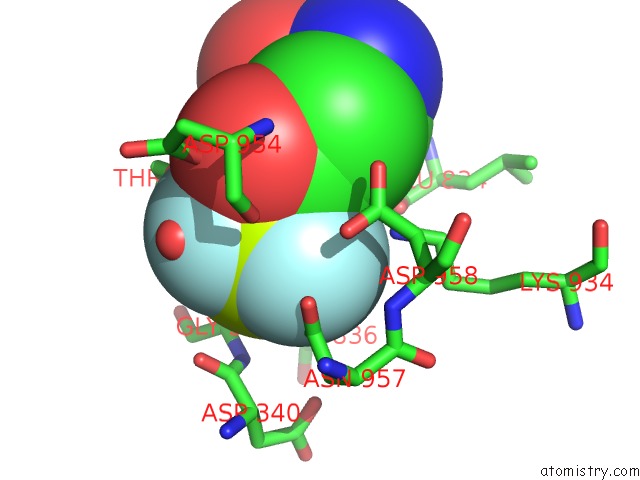
Mono view
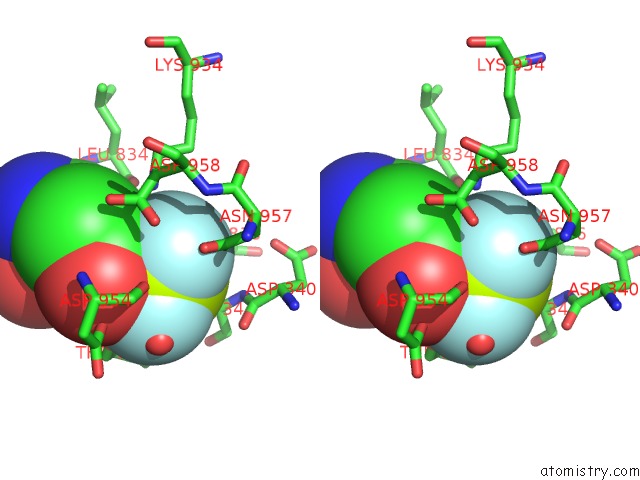
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Cryo-Em Structure of the Partially Activated DRS2P-CDC50P within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 6roi
Go back to
Fluorine binding site 2 out
of 3 in the Cryo-Em Structure of the Partially Activated DRS2P-CDC50P
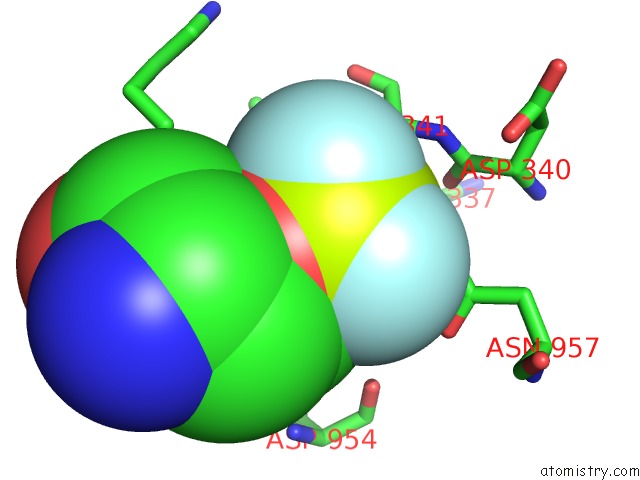
Mono view
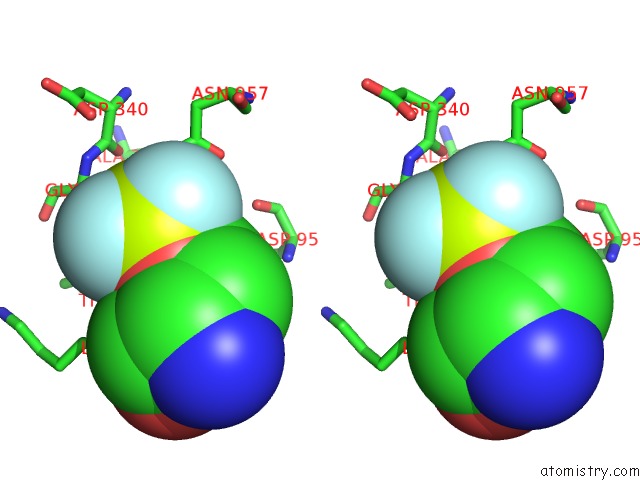
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Cryo-Em Structure of the Partially Activated DRS2P-CDC50P within 5.0Å range:
|
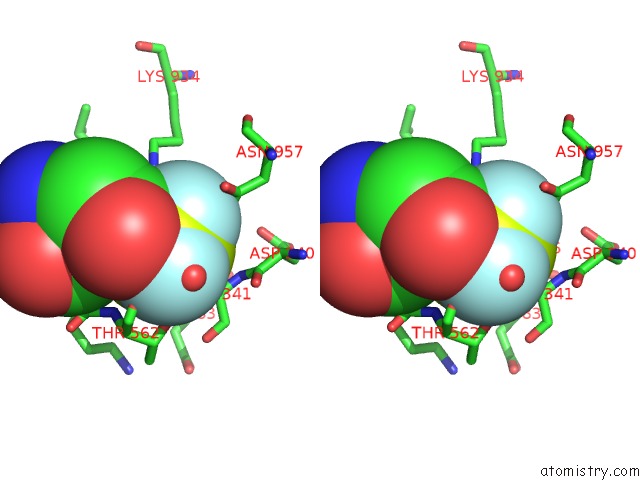
Fluorine binding site 3 out of 3 in 6roi
Go back to
Fluorine binding site 3 out
of 3 in the Cryo-Em Structure of the Partially Activated DRS2P-CDC50P
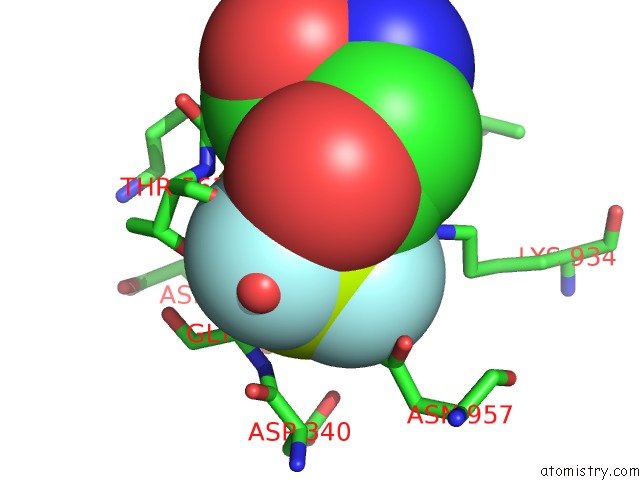
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Cryo-Em Structure of the Partially Activated DRS2P-CDC50P within 5.0Å range:
|
Reference:
M.Timcenko,
J.A.Lyons,
D.Januliene,
J.J.Ulstrup,
T.Dieudonne,
C.Montigny,
M.R.Ash,
J.L.Karlsen,
T.Boesen,
W.Kuhlbrandt,
G.Lenoir,
A.Moeller,
P.Nissen.
Structure and Autoregulation of A P4-Atpase Lipid Flippase. Nature V. 571 366 2019.
ISSN: ESSN 1476-4687
PubMed: 31243363
DOI: 10.1038/S41586-019-1344-7
Page generated: Tue Jul 15 15:23:17 2025
ISSN: ESSN 1476-4687
PubMed: 31243363
DOI: 10.1038/S41586-019-1344-7
Last articles
K in 9IS7K in 9HKX
K in 9HKY
K in 9HKW
K in 9HFO
K in 9HFN
K in 9HI3
K in 9HFM
K in 9HAC
K in 9GXH