Fluorine »
PDB 6yhd-6z4d »
6yuf »
Fluorine in PDB 6yuf: Cohesin Complex with Loader Gripping Dna
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Cohesin Complex with Loader Gripping Dna
(pdb code 6yuf). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 6 binding sites of Fluorine where determined in the Cohesin Complex with Loader Gripping Dna, PDB code: 6yuf:
Jump to Fluorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Fluorine where determined in the Cohesin Complex with Loader Gripping Dna, PDB code: 6yuf:
Jump to Fluorine binding site number: 1; 2; 3; 4; 5; 6;
Fluorine binding site 1 out of 6 in 6yuf
Go back to
Fluorine binding site 1 out
of 6 in the Cohesin Complex with Loader Gripping Dna
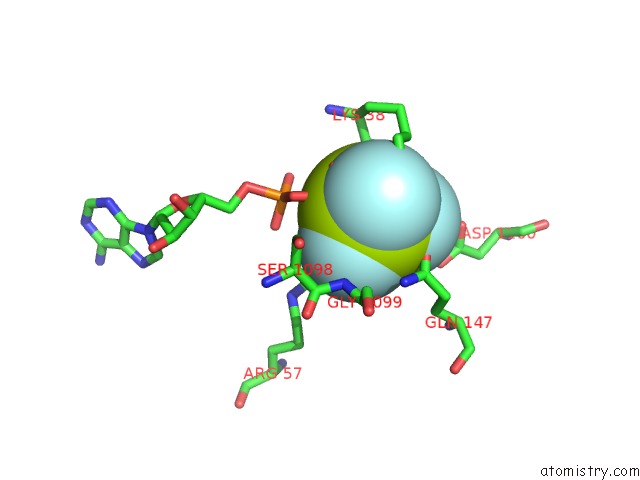
Mono view
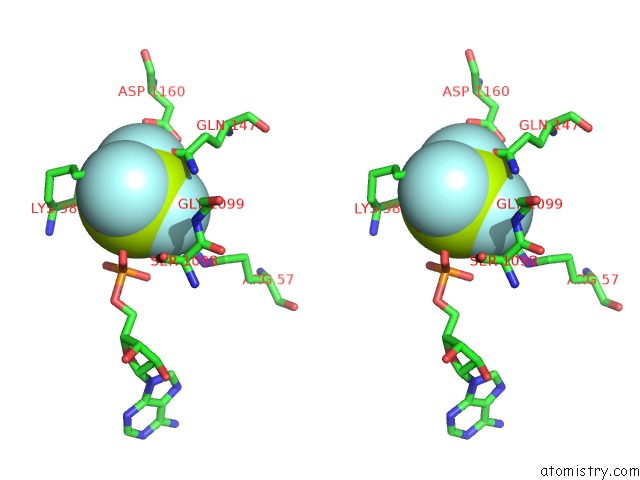
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Cohesin Complex with Loader Gripping Dna within 5.0Å range:
|
Fluorine binding site 2 out of 6 in 6yuf
Go back to
Fluorine binding site 2 out
of 6 in the Cohesin Complex with Loader Gripping Dna
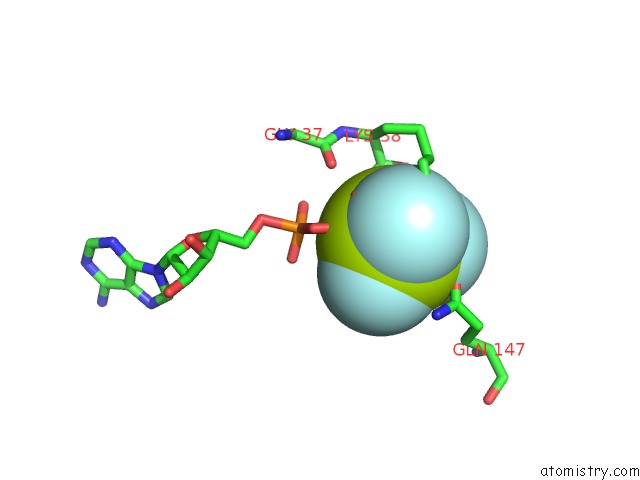
Mono view
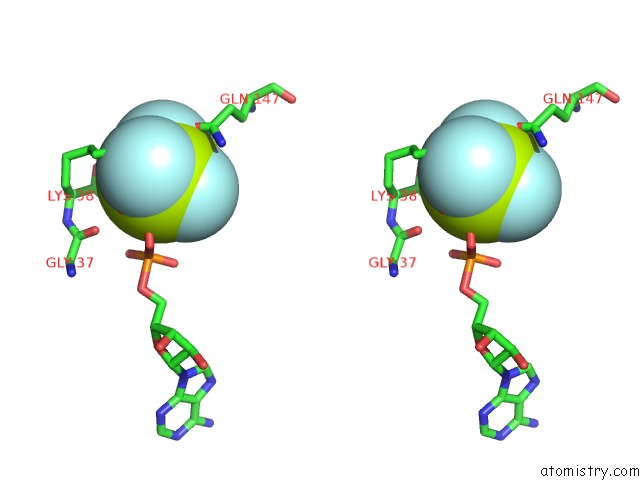
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Cohesin Complex with Loader Gripping Dna within 5.0Å range:
|
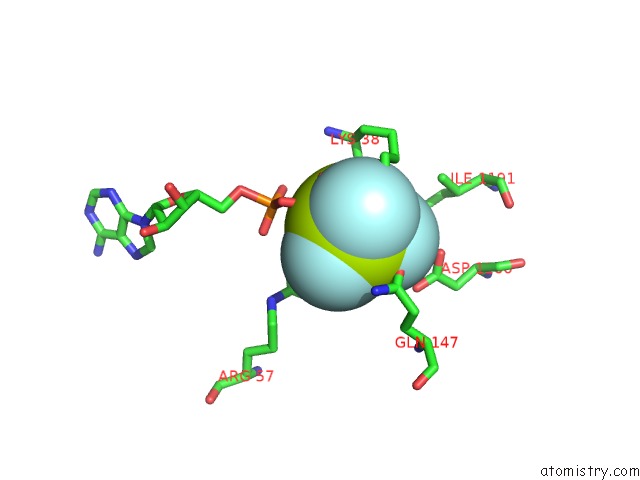
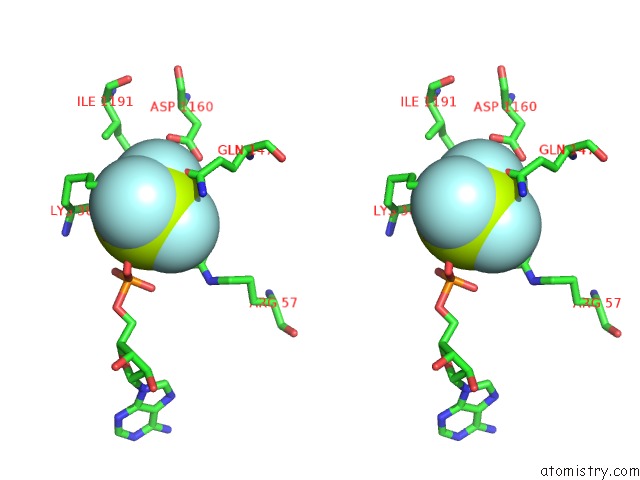
Fluorine binding site 3 out of 6 in 6yuf
Go back to
Fluorine binding site 3 out
of 6 in the Cohesin Complex with Loader Gripping Dna
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Cohesin Complex with Loader Gripping Dna within 5.0Å range:
|
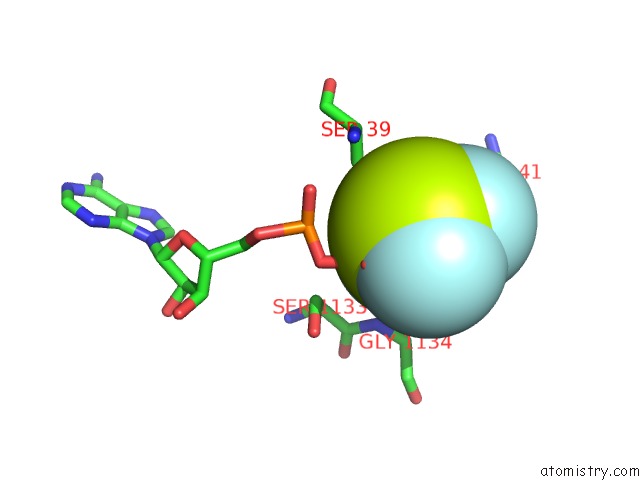
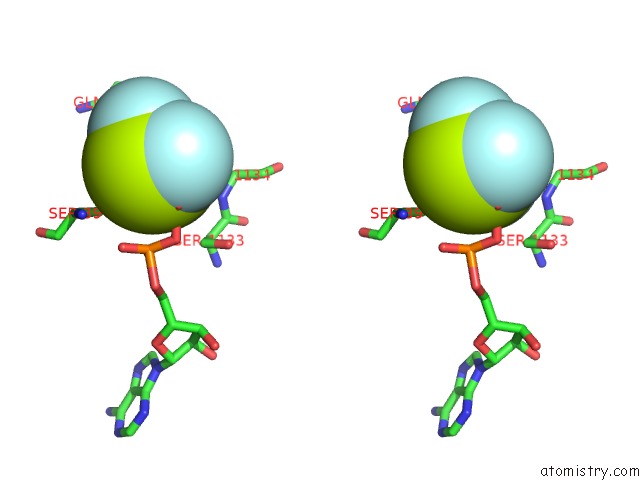
Fluorine binding site 4 out of 6 in 6yuf
Go back to
Fluorine binding site 4 out
of 6 in the Cohesin Complex with Loader Gripping Dna
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 4 of Cohesin Complex with Loader Gripping Dna within 5.0Å range:
|
Fluorine binding site 5 out of 6 in 6yuf
Go back to
Fluorine binding site 5 out
of 6 in the Cohesin Complex with Loader Gripping Dna
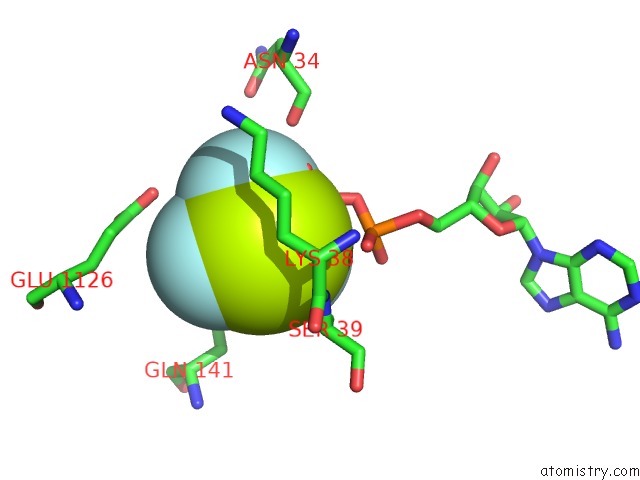
Mono view
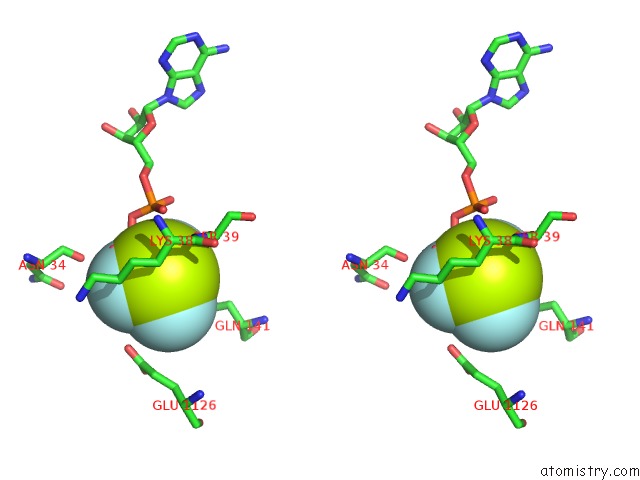
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 5 of Cohesin Complex with Loader Gripping Dna within 5.0Å range:
|
Fluorine binding site 6 out of 6 in 6yuf
Go back to
Fluorine binding site 6 out
of 6 in the Cohesin Complex with Loader Gripping Dna
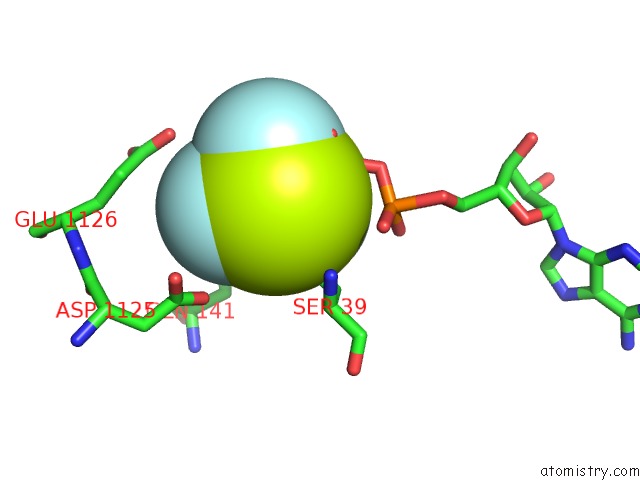
Mono view
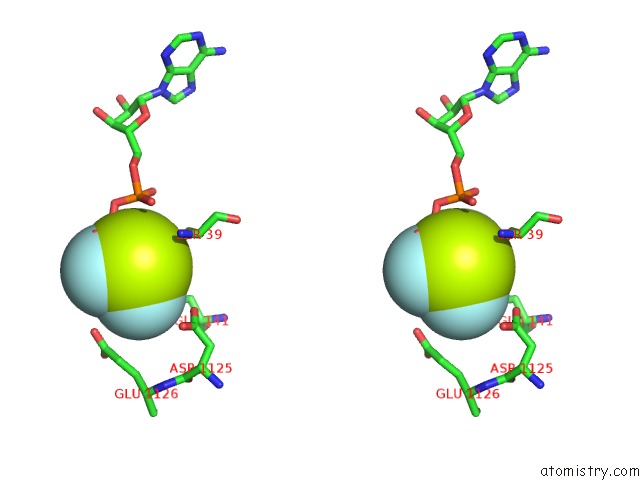
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 6 of Cohesin Complex with Loader Gripping Dna within 5.0Å range:
|
Reference:
T.L.Higashi,
P.Eickhoff,
J.S.Sousa,
J.Locke,
A.Nans,
H.R.Flynn,
A.P.Snijders,
G.Papageorgiou,
N.O'reilly,
Z.A.Chen,
F.J.O'reilly,
J.Rappsilber,
A.Costa,
F.Uhlmann.
A Structure-Based Mechanism For Dna Entry Into the Cohesin Ring. Mol.Cell V. 79 917 2020.
ISSN: ISSN 1097-2765
PubMed: 32755595
DOI: 10.1016/J.MOLCEL.2020.07.013
Page generated: Tue Jul 15 17:56:41 2025
ISSN: ISSN 1097-2765
PubMed: 32755595
DOI: 10.1016/J.MOLCEL.2020.07.013
Last articles
Mg in 6HVYMg in 6HW3
Mg in 6HW0
Mg in 6HW1
Mg in 6HVX
Mg in 6HQB
Mg in 6HVW
Mg in 6HVV
Mg in 6HVU
Mg in 6HVT