Fluorine »
PDB 7f97-7fmn »
7flb »
Fluorine in PDB 7flb: Pandda Analysis Group Deposition -- AAR2/Rnaseh in Complex with Fragment P05A10 From the F2X-Universal Library
Protein crystallography data
The structure of Pandda Analysis Group Deposition -- AAR2/Rnaseh in Complex with Fragment P05A10 From the F2X-Universal Library, PDB code: 7flb
was solved by
T.Barthel,
J.Wollenhaupt,
G.M.A.Lima,
M.C.Wahl,
M.S.Weiss,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.04 / 1.57 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 89.592, 82.079, 93.849, 90, 108.34, 90 |
| R / Rfree (%) | 22 / 25.4 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Pandda Analysis Group Deposition -- AAR2/Rnaseh in Complex with Fragment P05A10 From the F2X-Universal Library
(pdb code 7flb). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Pandda Analysis Group Deposition -- AAR2/Rnaseh in Complex with Fragment P05A10 From the F2X-Universal Library, PDB code: 7flb:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Pandda Analysis Group Deposition -- AAR2/Rnaseh in Complex with Fragment P05A10 From the F2X-Universal Library, PDB code: 7flb:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 7flb
Go back to
Fluorine binding site 1 out
of 2 in the Pandda Analysis Group Deposition -- AAR2/Rnaseh in Complex with Fragment P05A10 From the F2X-Universal Library
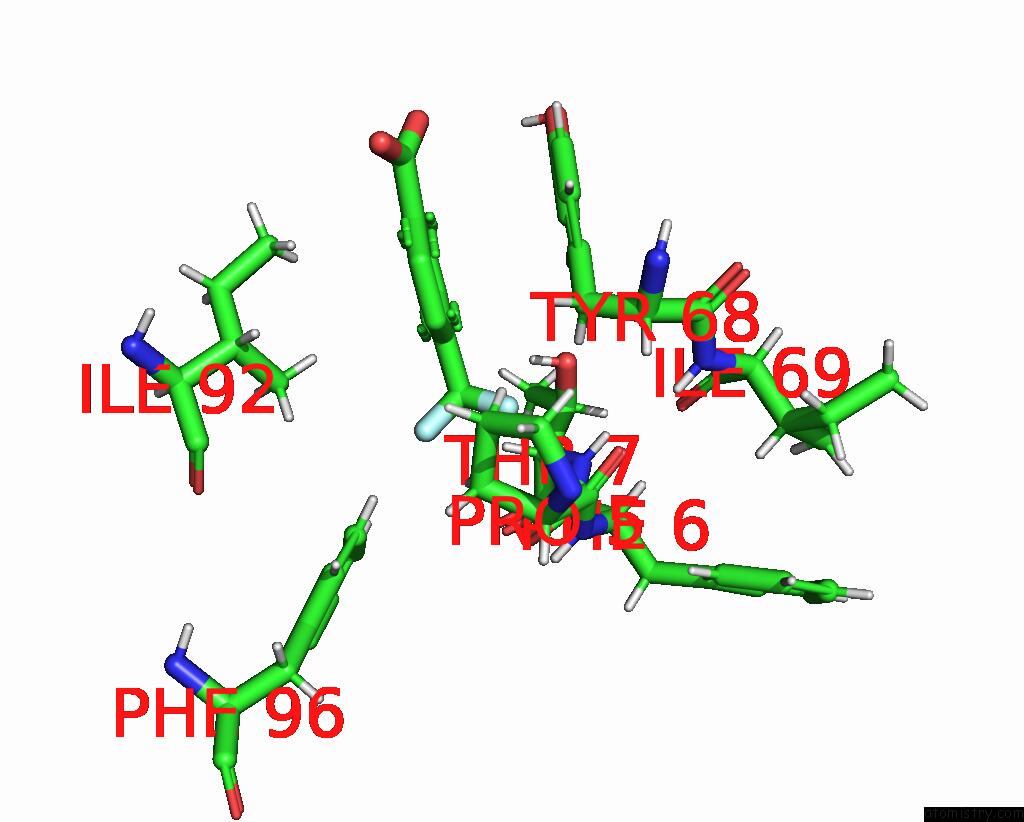
Mono view
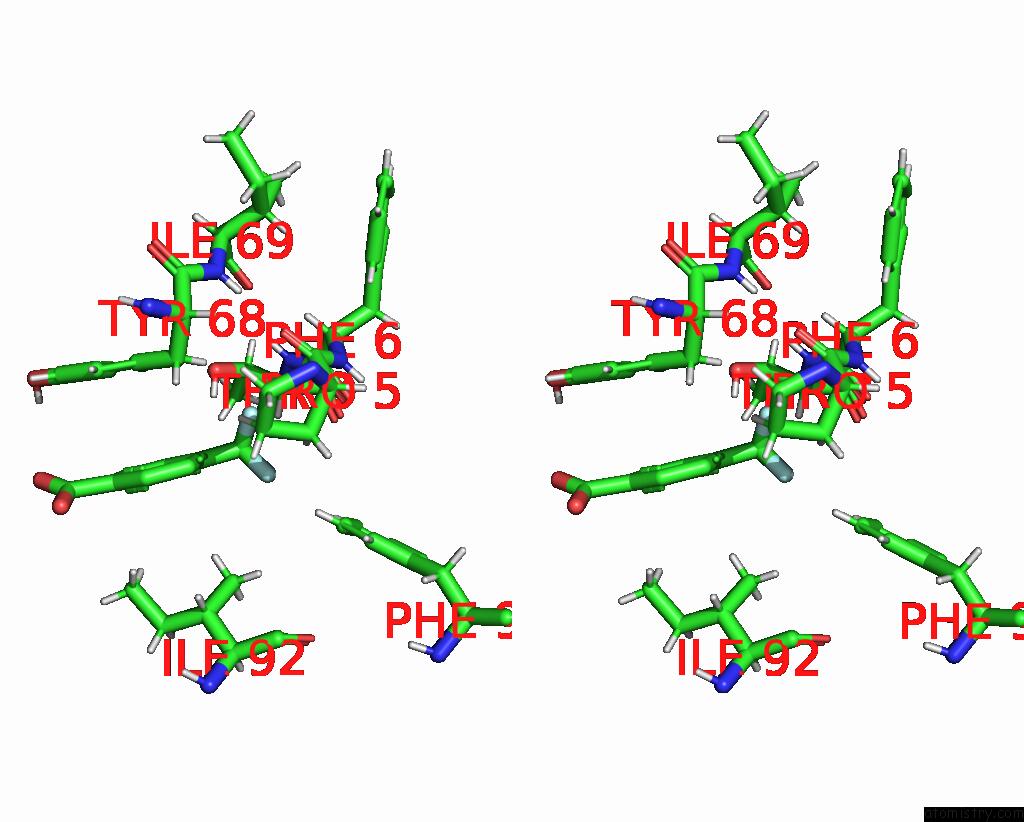
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Pandda Analysis Group Deposition -- AAR2/Rnaseh in Complex with Fragment P05A10 From the F2X-Universal Library within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 7flb
Go back to
Fluorine binding site 2 out
of 2 in the Pandda Analysis Group Deposition -- AAR2/Rnaseh in Complex with Fragment P05A10 From the F2X-Universal Library
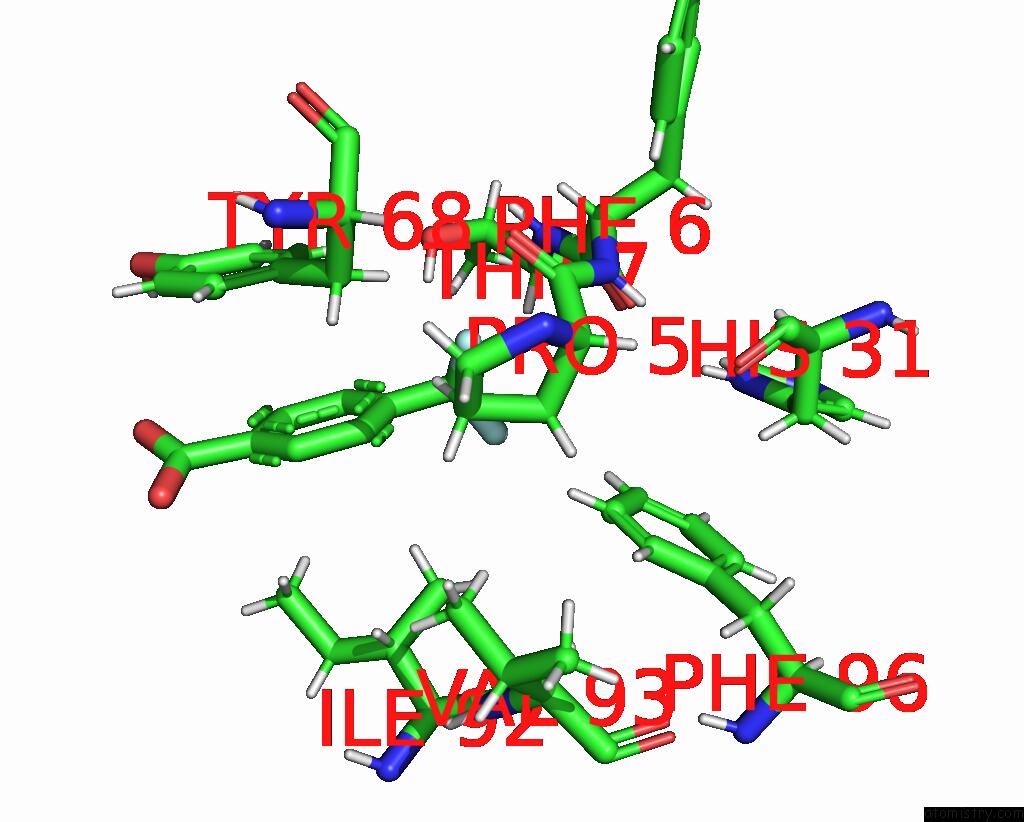
Mono view
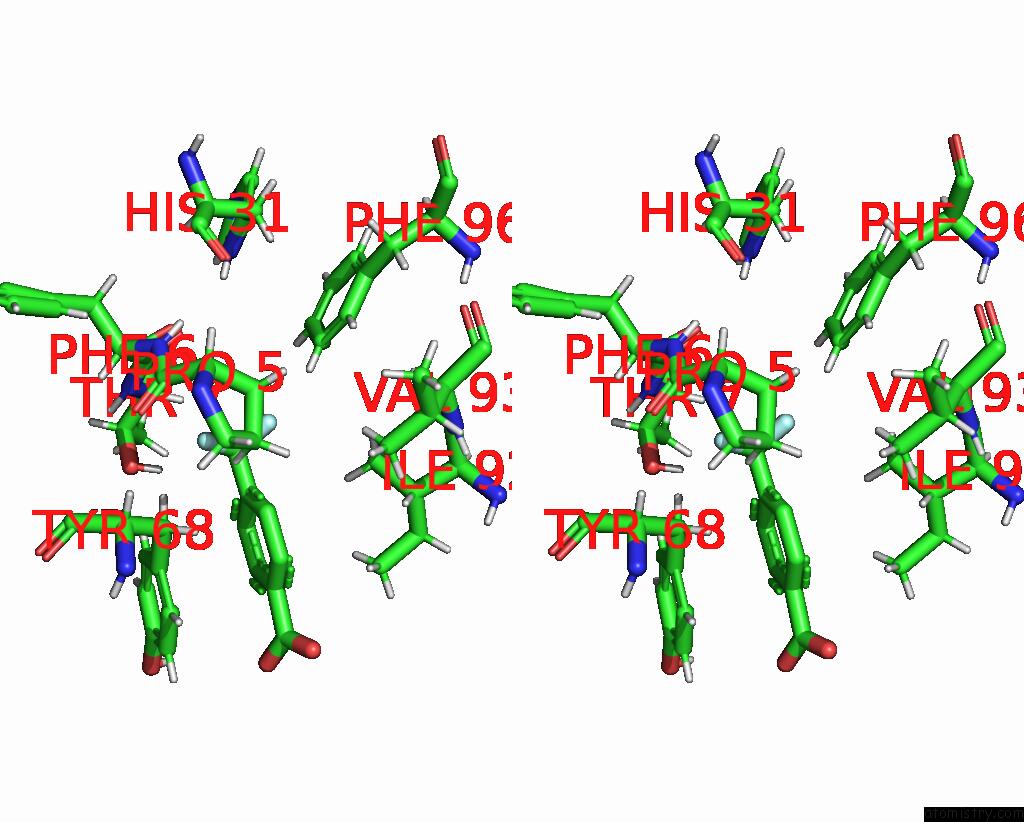
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Pandda Analysis Group Deposition -- AAR2/Rnaseh in Complex with Fragment P05A10 From the F2X-Universal Library within 5.0Å range:
|
Reference:
T.Barthel,
J.Wollenhaupt,
G.M.A.Lima,
M.C.Wahl,
M.S.Weiss.
Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites. J.Med.Chem. V. 65 14630 2022.
ISSN: ISSN 0022-2623
PubMed: 36260741
DOI: 10.1021/ACS.JMEDCHEM.2C01165
Page generated: Tue Jul 15 19:33:24 2025
ISSN: ISSN 0022-2623
PubMed: 36260741
DOI: 10.1021/ACS.JMEDCHEM.2C01165
Last articles
I in 2GS7I in 2H0S
I in 2GJM
I in 2GSQ
I in 2GGQ
I in 2FWZ
I in 2G19
I in 2G1M
I in 2DGM
I in 2FWE