Fluorine »
PDB 8fm3-8fy1 »
8fw5 »
Fluorine in PDB 8fw5: Chimeric HSGATOR1-Spgtr-Splam Complex
Other elements in 8fw5:
The structure of Chimeric HSGATOR1-Spgtr-Splam Complex also contains other interesting chemical elements:
| Aluminium | (Al) | 1 atom |
| Magnesium | (Mg) | 1 atom |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Chimeric HSGATOR1-Spgtr-Splam Complex
(pdb code 8fw5). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Chimeric HSGATOR1-Spgtr-Splam Complex, PDB code: 8fw5:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Chimeric HSGATOR1-Spgtr-Splam Complex, PDB code: 8fw5:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 8fw5
Go back to
Fluorine binding site 1 out
of 3 in the Chimeric HSGATOR1-Spgtr-Splam Complex

Mono view
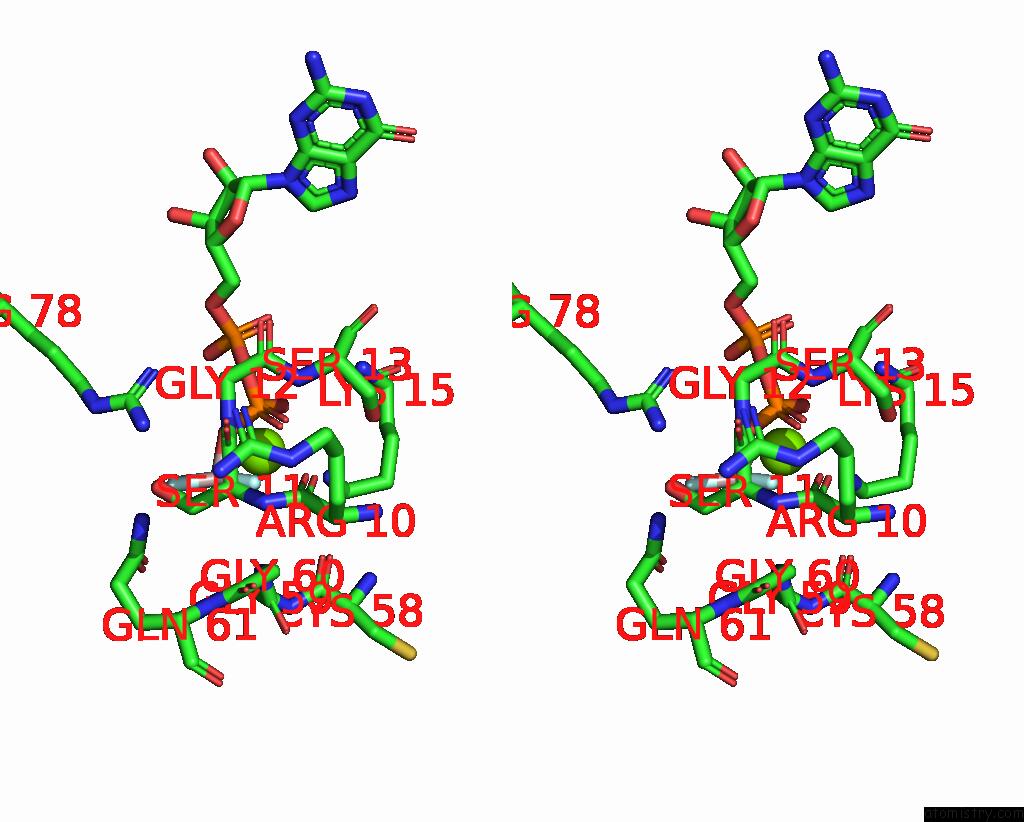
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Chimeric HSGATOR1-Spgtr-Splam Complex within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 8fw5
Go back to
Fluorine binding site 2 out
of 3 in the Chimeric HSGATOR1-Spgtr-Splam Complex
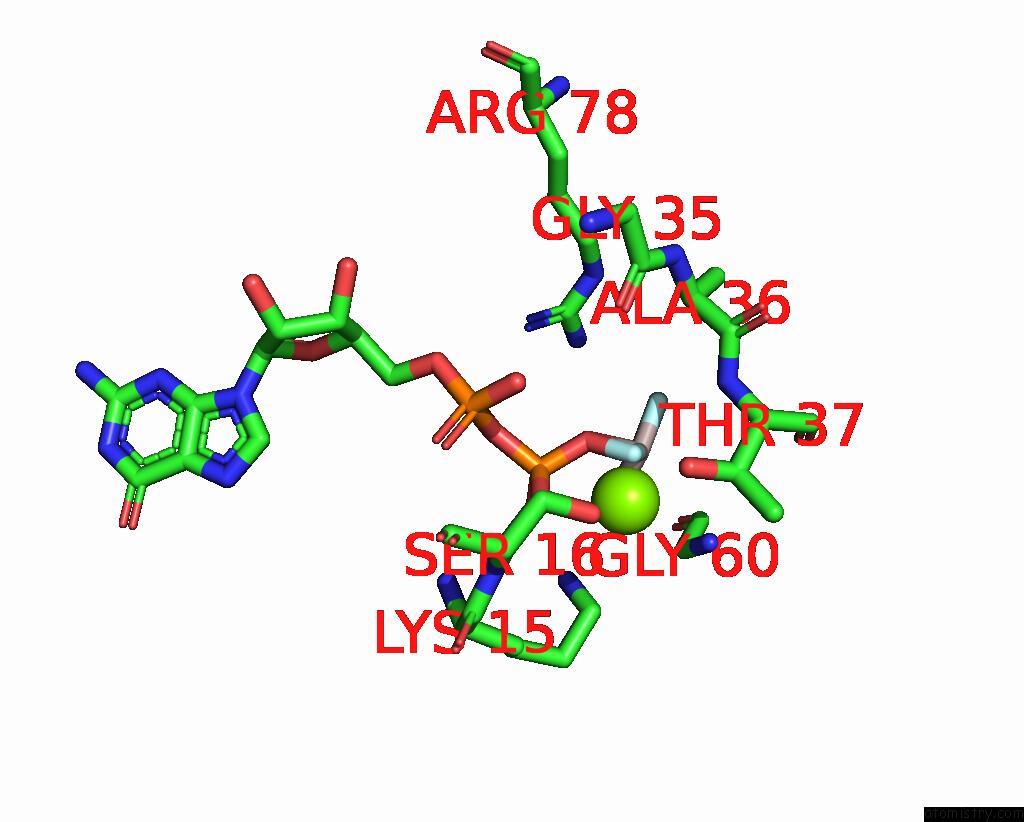
Mono view
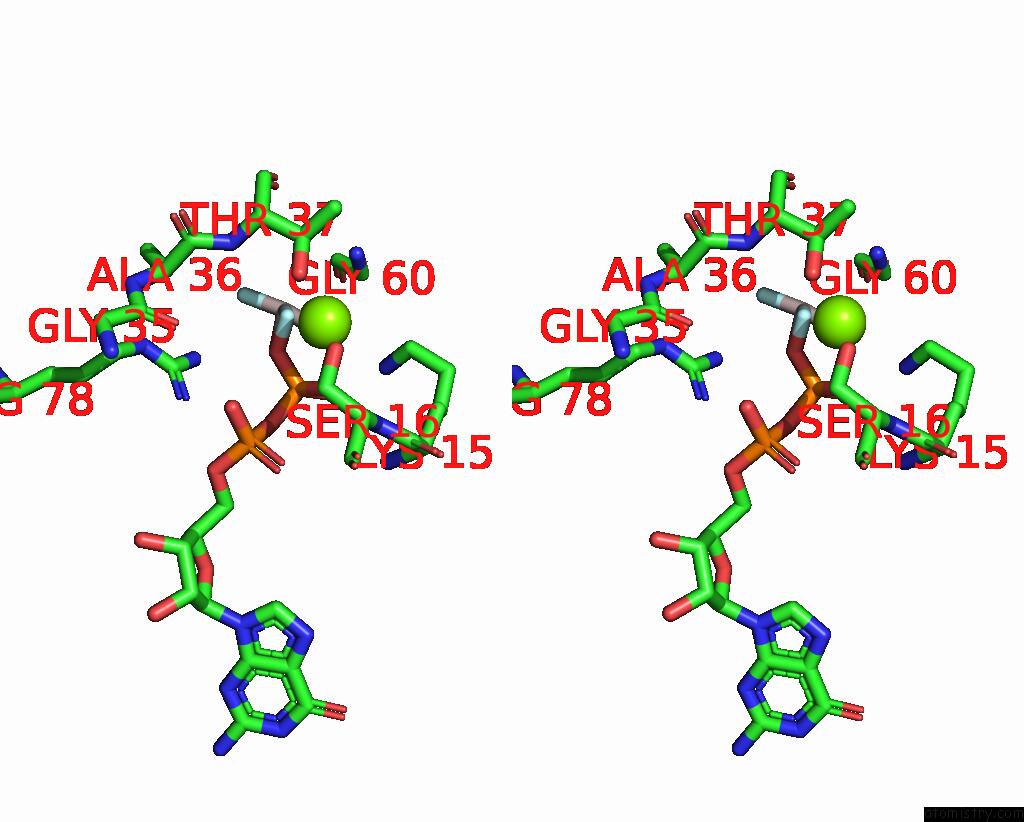
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Chimeric HSGATOR1-Spgtr-Splam Complex within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 8fw5
Go back to
Fluorine binding site 3 out
of 3 in the Chimeric HSGATOR1-Spgtr-Splam Complex
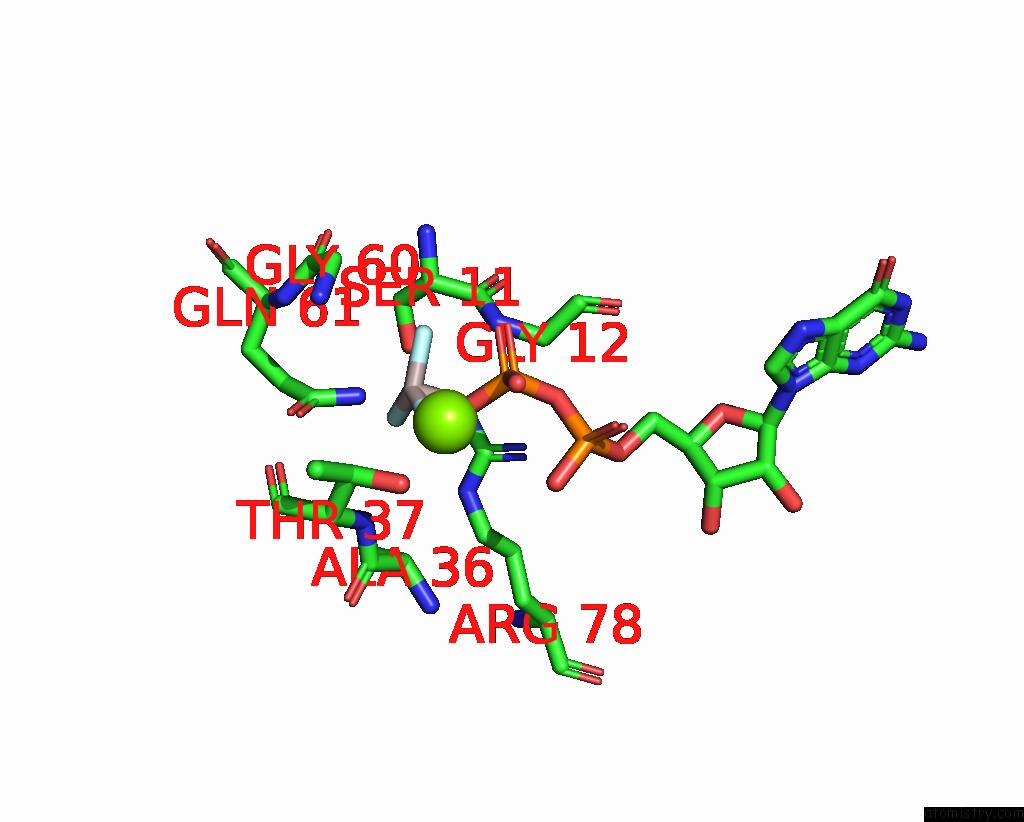
Mono view
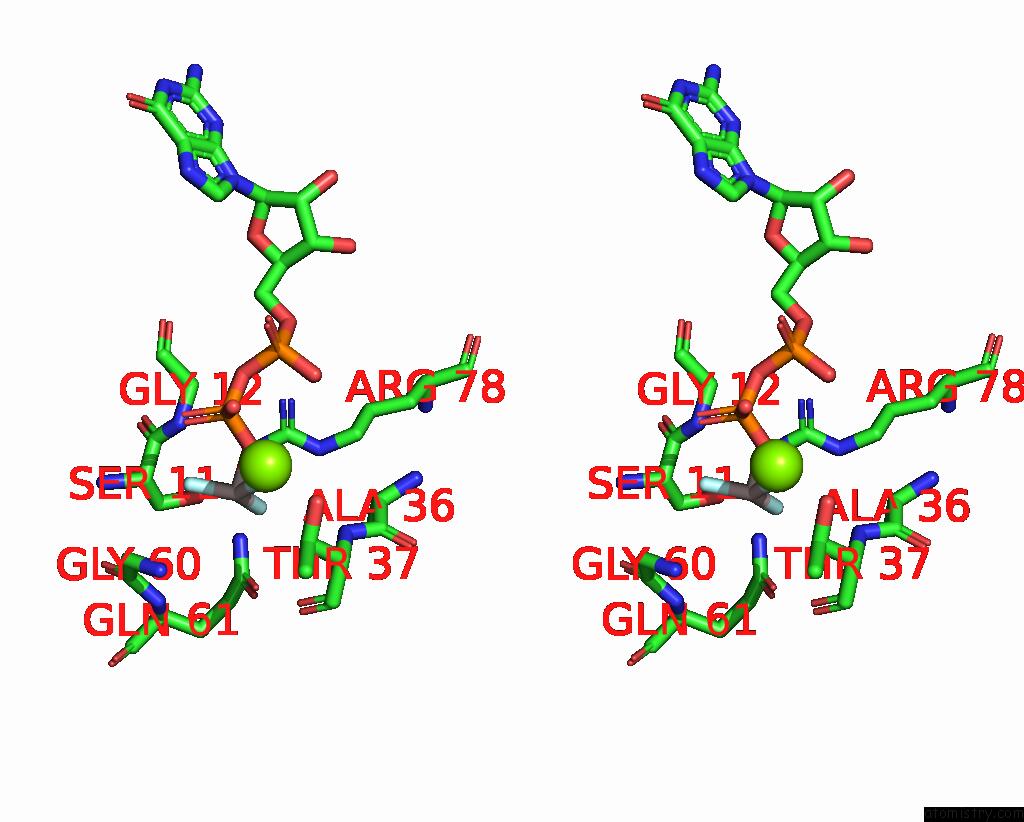
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Chimeric HSGATOR1-Spgtr-Splam Complex within 5.0Å range:
|
Reference:
S.D.Tettoni,
S.B.Egri,
D.D.Doxsey,
K.Veinotte,
C.Ouch,
J.Y.Chang,
K.Song,
C.Xu,
K.Shen.
Structure of the Schizosaccharomyces Pombe Gtr-Lam Complex Reveals Evolutionary Divergence of MTORC1-Dependent Amino Acid Sensing. Structure 2023.
ISSN: ISSN 0969-2126
PubMed: 37453417
DOI: 10.1016/J.STR.2023.06.012
Page generated: Wed Jul 16 04:47:50 2025
ISSN: ISSN 0969-2126
PubMed: 37453417
DOI: 10.1016/J.STR.2023.06.012
Last articles
Mn in 9LJUMn in 9LJW
Mn in 9LJS
Mn in 9LJR
Mn in 9LJT
Mn in 9LJV
Mg in 9UA2
Mg in 9R96
Mg in 9VM1
Mg in 9P01