Fluorine »
PDB 8g63-8gky »
8gfu »
Fluorine in PDB 8gfu: Room Temperature X-Ray Structure of Truncated Sars-Cov-2 Main Protease C145A Mutant, Residues 1-304, in Complex with Nirmatrelvir (Nmv)
Protein crystallography data
The structure of Room Temperature X-Ray Structure of Truncated Sars-Cov-2 Main Protease C145A Mutant, Residues 1-304, in Complex with Nirmatrelvir (Nmv), PDB code: 8gfu
was solved by
A.Kovalevsky,
L.Coates,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.25 / 1.80 |
| Space group | I 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.223, 81.711, 91.702, 90, 95.39, 90 |
| R / Rfree (%) | 16.6 / 19.3 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Room Temperature X-Ray Structure of Truncated Sars-Cov-2 Main Protease C145A Mutant, Residues 1-304, in Complex with Nirmatrelvir (Nmv)
(pdb code 8gfu). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Room Temperature X-Ray Structure of Truncated Sars-Cov-2 Main Protease C145A Mutant, Residues 1-304, in Complex with Nirmatrelvir (Nmv), PDB code: 8gfu:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Room Temperature X-Ray Structure of Truncated Sars-Cov-2 Main Protease C145A Mutant, Residues 1-304, in Complex with Nirmatrelvir (Nmv), PDB code: 8gfu:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 8gfu
Go back to
Fluorine binding site 1 out
of 3 in the Room Temperature X-Ray Structure of Truncated Sars-Cov-2 Main Protease C145A Mutant, Residues 1-304, in Complex with Nirmatrelvir (Nmv)
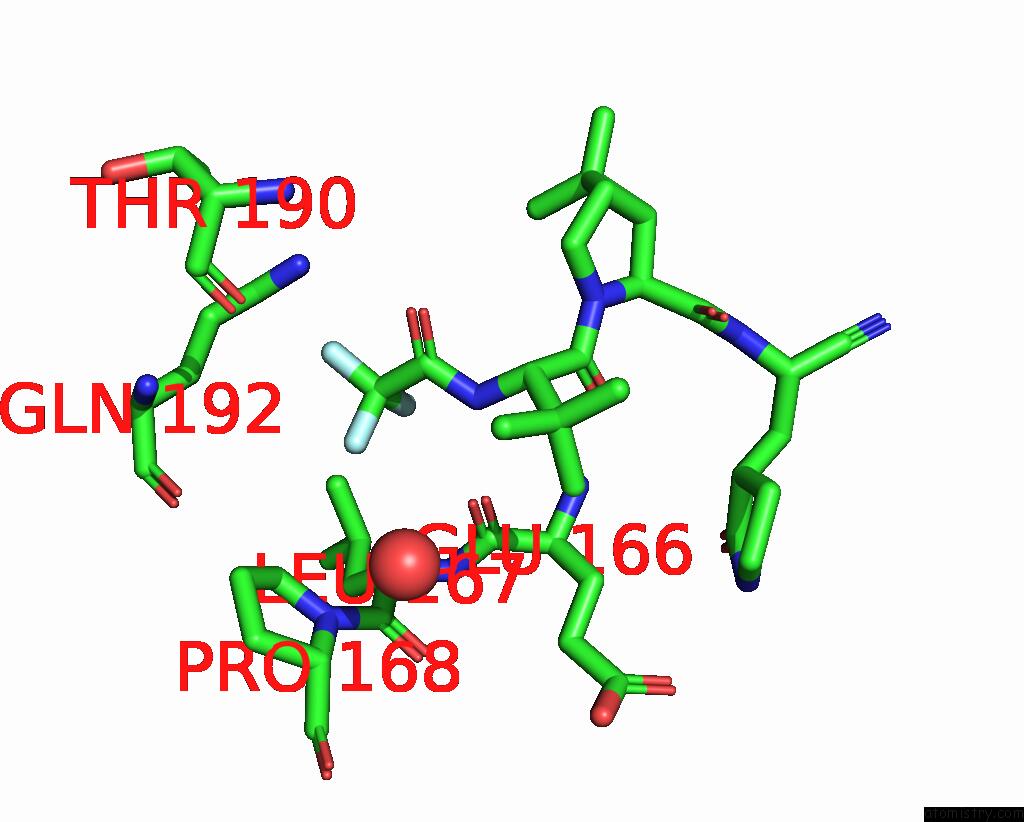
Mono view
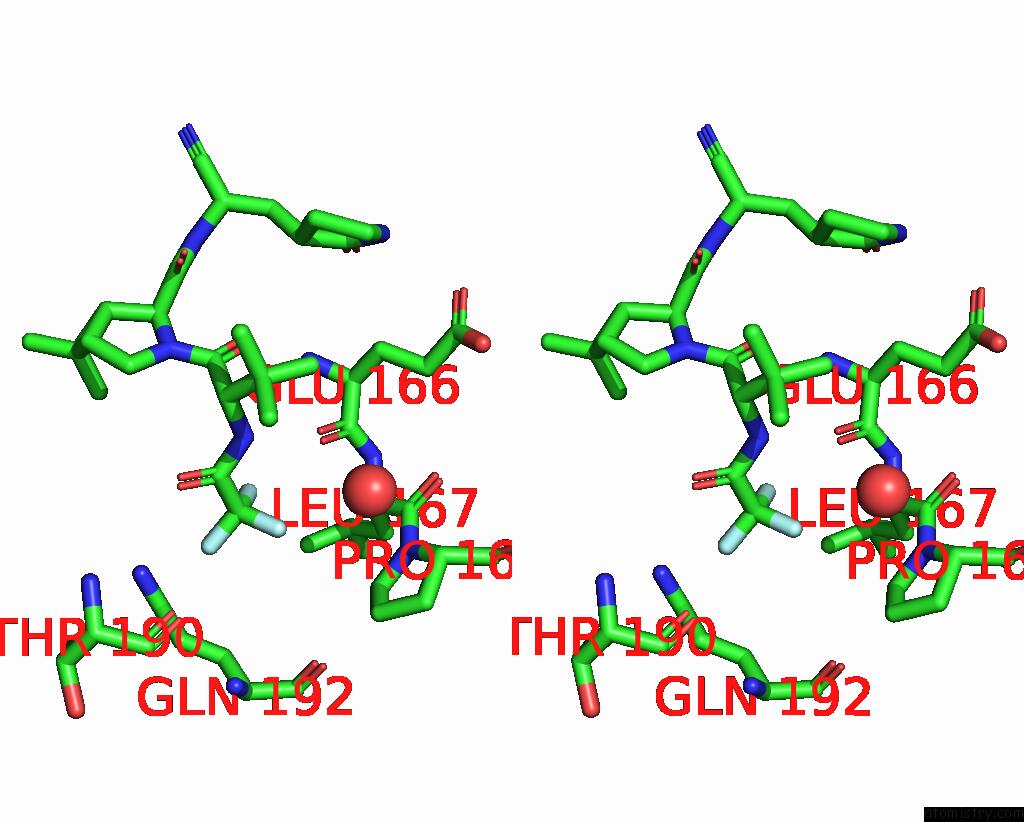
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Room Temperature X-Ray Structure of Truncated Sars-Cov-2 Main Protease C145A Mutant, Residues 1-304, in Complex with Nirmatrelvir (Nmv) within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 8gfu
Go back to
Fluorine binding site 2 out
of 3 in the Room Temperature X-Ray Structure of Truncated Sars-Cov-2 Main Protease C145A Mutant, Residues 1-304, in Complex with Nirmatrelvir (Nmv)

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Room Temperature X-Ray Structure of Truncated Sars-Cov-2 Main Protease C145A Mutant, Residues 1-304, in Complex with Nirmatrelvir (Nmv) within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 8gfu
Go back to
Fluorine binding site 3 out
of 3 in the Room Temperature X-Ray Structure of Truncated Sars-Cov-2 Main Protease C145A Mutant, Residues 1-304, in Complex with Nirmatrelvir (Nmv)
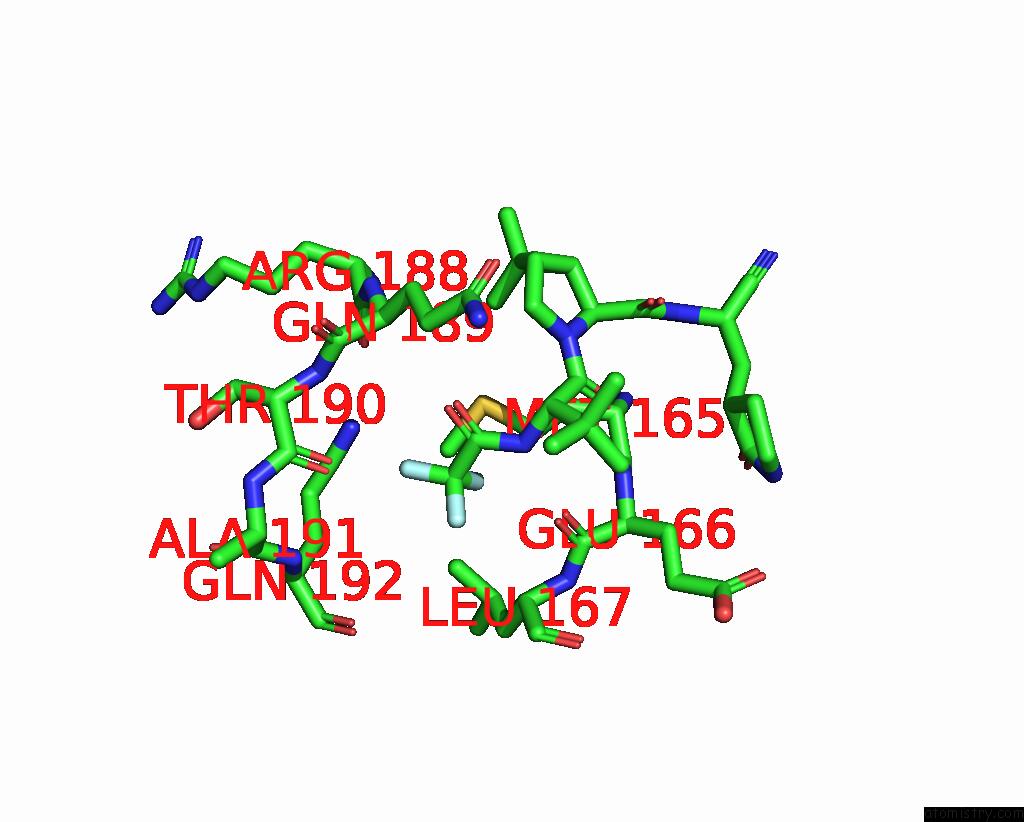
Mono view
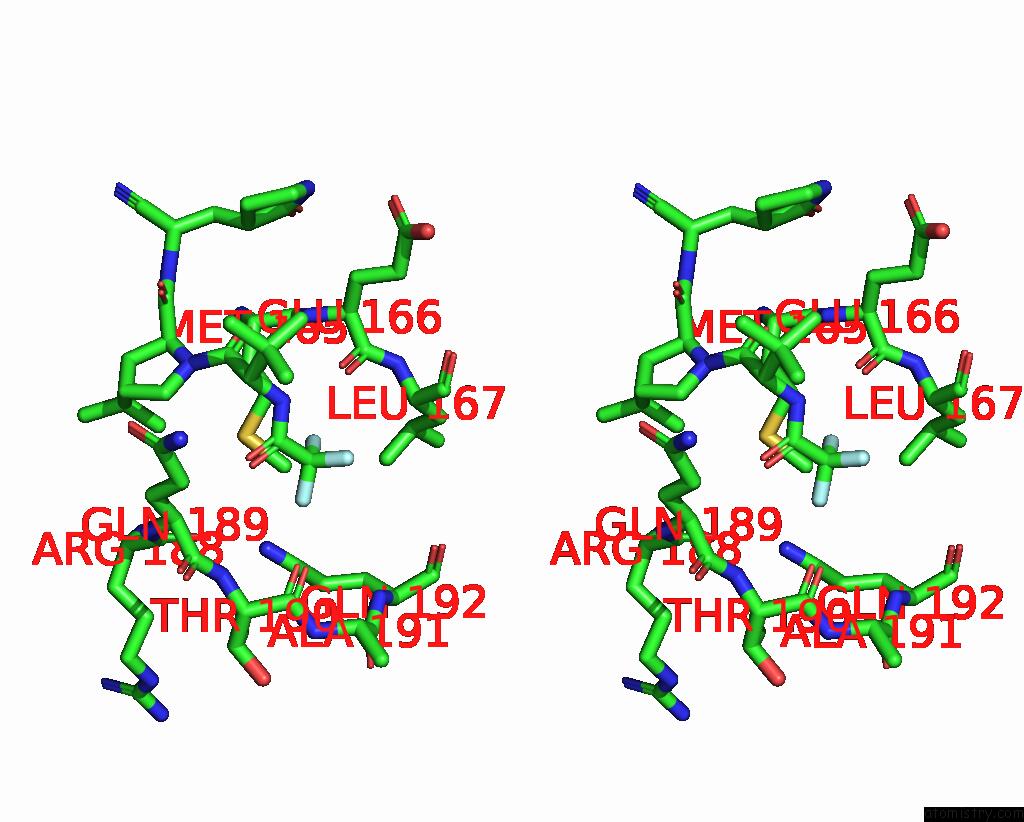
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Room Temperature X-Ray Structure of Truncated Sars-Cov-2 Main Protease C145A Mutant, Residues 1-304, in Complex with Nirmatrelvir (Nmv) within 5.0Å range:
|
Reference:
A.Kovalevsky,
A.Aniana,
L.Coates,
P.V.Bonnesen,
N.T.Nashed,
J.M.Louis.
Contribution of the Catalytic Dyad of Sars-Cov-2 Main Protease to Binding Covalent and Noncovalent Inhibitors. J.Biol.Chem. V. 299 04886 2023.
ISSN: ESSN 1083-351X
PubMed: 37271339
DOI: 10.1016/J.JBC.2023.104886
Page generated: Wed Jul 16 05:00:25 2025
ISSN: ESSN 1083-351X
PubMed: 37271339
DOI: 10.1016/J.JBC.2023.104886
Last articles
Mn in 5RAAMn in 5OMX
Mn in 5OXJ
Mn in 5R7X
Mn in 5OX6
Mn in 5OX5
Mn in 5OR6
Mn in 5OVO
Mn in 5OR2
Mn in 5ONW