Fluorine »
PDB 8hyp-8ijw »
8ijv »
Fluorine in PDB 8ijv: Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-02
Other elements in 8ijv:
The structure of Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-02 also contains other interesting chemical elements:
| Chlorine | (Cl) | 4 atoms |
| Magnesium | (Mg) | 1 atom |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-02
(pdb code 8ijv). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-02, PDB code: 8ijv:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-02, PDB code: 8ijv:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 8ijv
Go back to
Fluorine binding site 1 out
of 3 in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-02
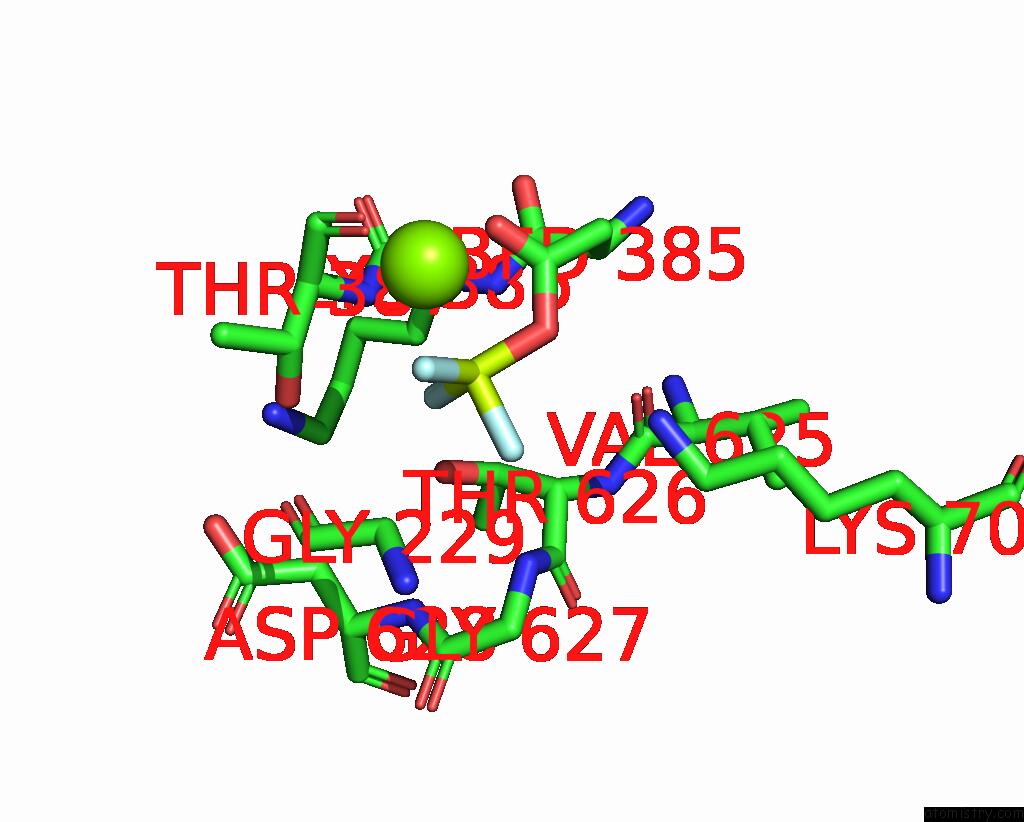
Mono view
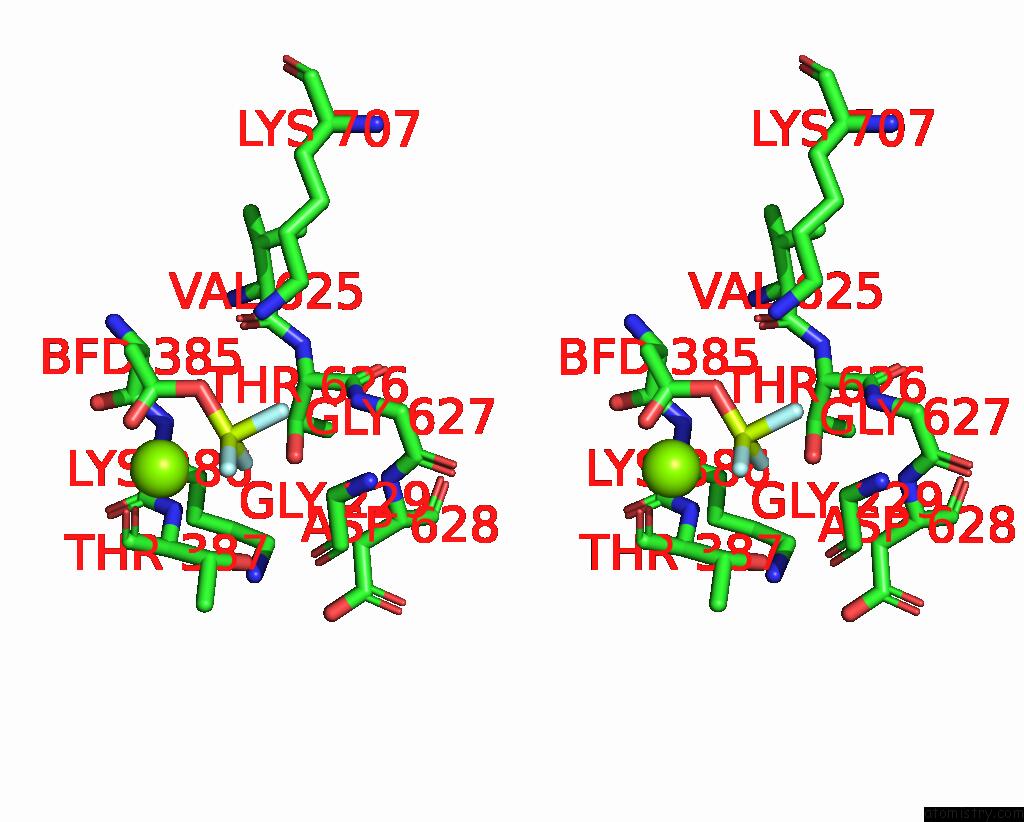
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-02 within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 8ijv
Go back to
Fluorine binding site 2 out
of 3 in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-02
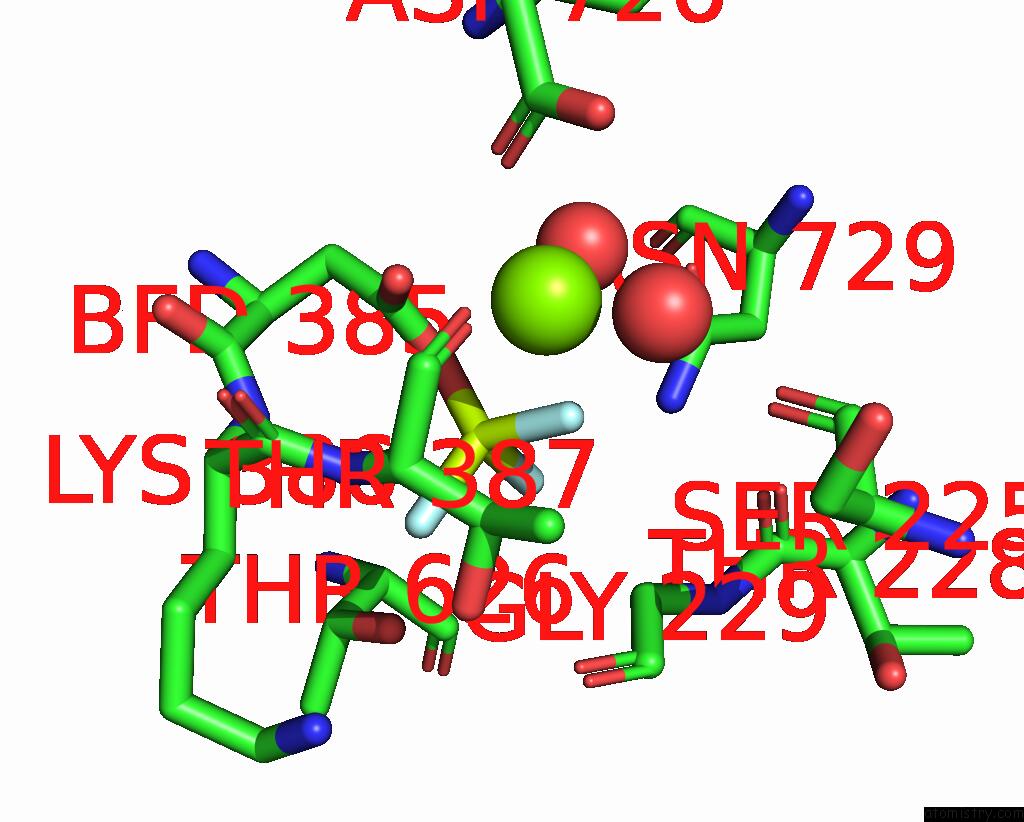
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-02 within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 8ijv
Go back to
Fluorine binding site 3 out
of 3 in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-02
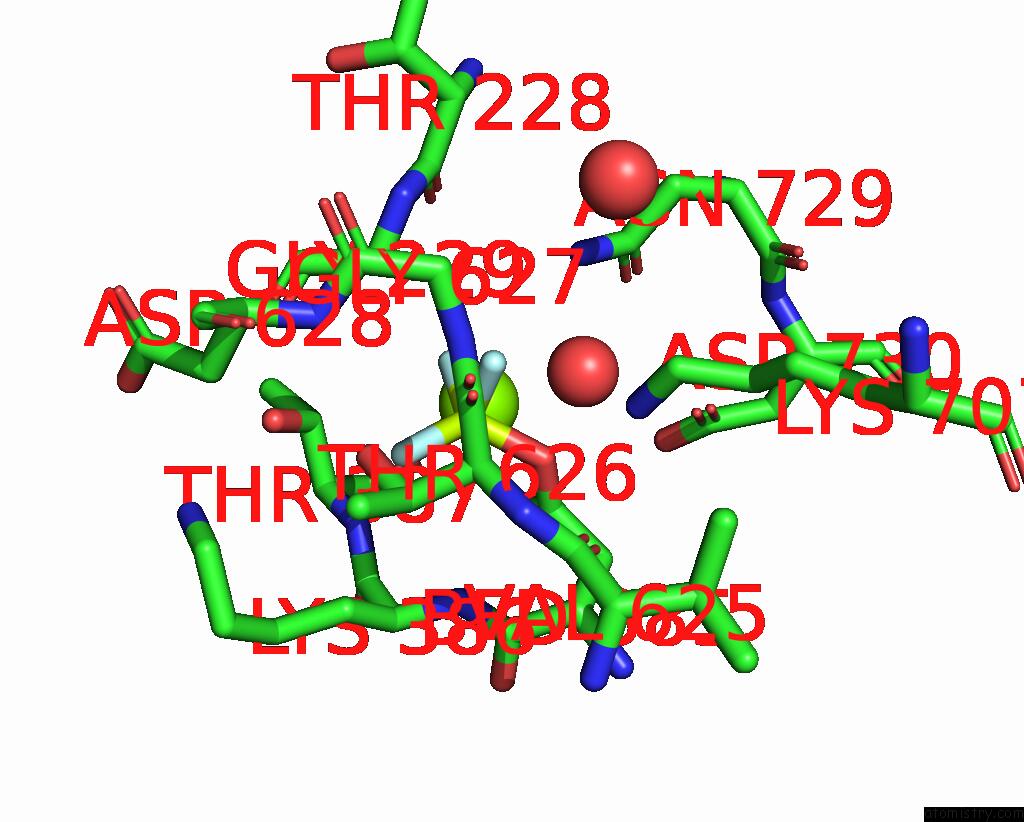
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-02 within 5.0Å range:
|
Reference:
K.Abe,
S.Yokoshima,
A.Yoshimori.
Deep Learning Driven De Novo Drug Design Based on Gastric Proton Pump Structures To Be Published.
Page generated: Wed Jul 16 05:38:36 2025
Last articles
K in 8S3EK in 8S4S
K in 8S1X
K in 8RUH
K in 8RUJ
K in 8RUI
K in 8RZ6
K in 8RMH
K in 8RS5
K in 8R6F