Fluorine »
PDB 8ijx-8jop »
8jmn »
Fluorine in PDB 8jmn: Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-21
Other elements in 8jmn:
The structure of Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-21 also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
| Chlorine | (Cl) | 1 atom |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-21
(pdb code 8jmn). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-21, PDB code: 8jmn:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-21, PDB code: 8jmn:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 8jmn
Go back to
Fluorine binding site 1 out
of 3 in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-21

Mono view
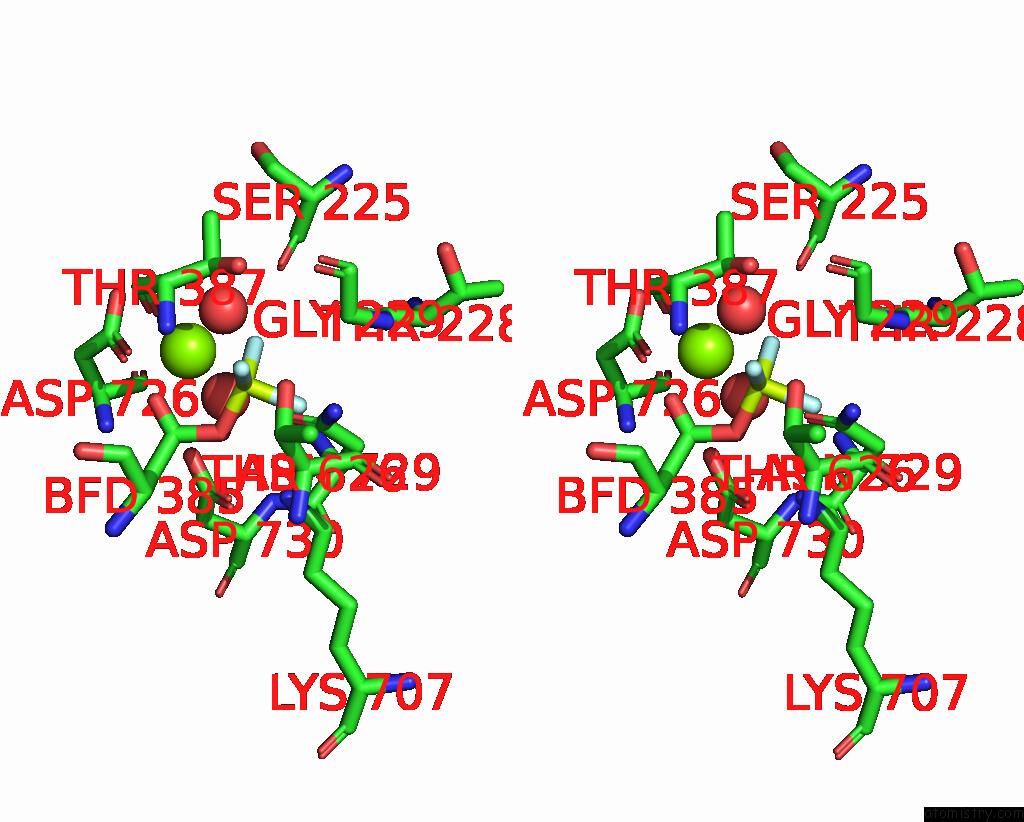
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-21 within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 8jmn
Go back to
Fluorine binding site 2 out
of 3 in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-21

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-21 within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 8jmn
Go back to
Fluorine binding site 3 out
of 3 in the Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-21
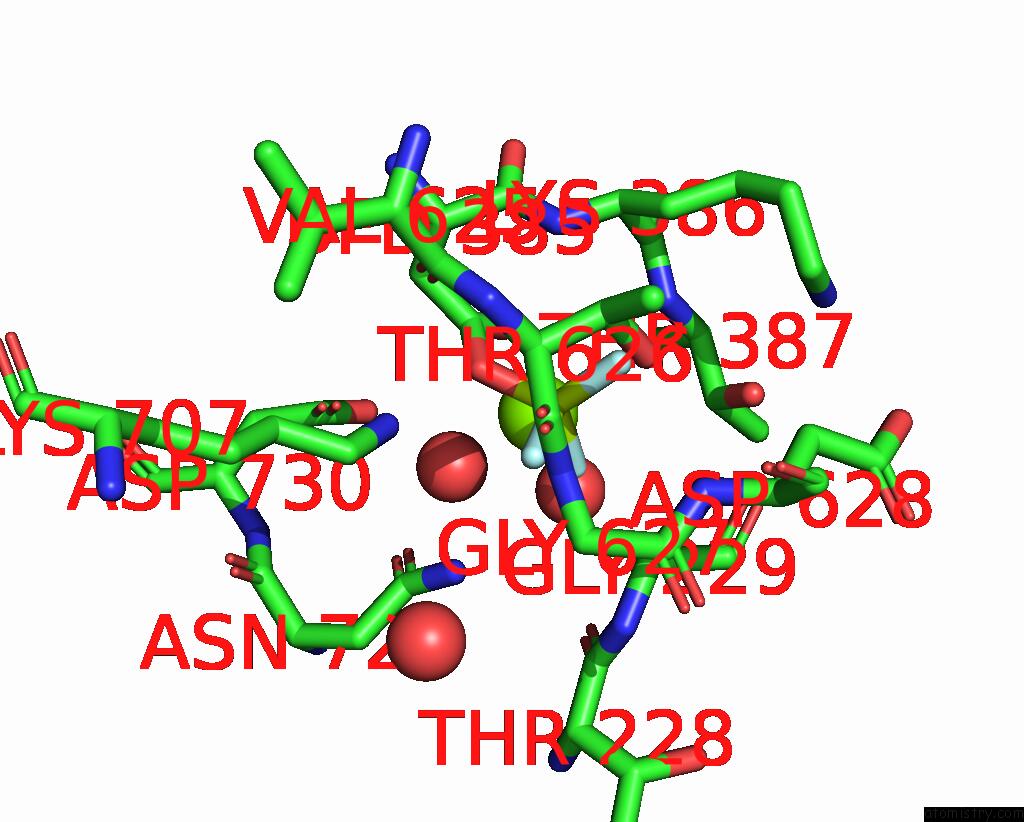
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Cryo-Em Structure of the Gastric Proton Pump with Bound Dq-21 within 5.0Å range:
|
Reference:
K.Abe,
S.Yokoshima,
A.Yoshimori.
Deep Learning Driven De Novo Drug Design Based on Gastric Proton Pump Structures To Be Published.
Page generated: Wed Jul 16 05:48:21 2025
Last articles
Mg in 5SE8Mg in 5SE7
Mg in 5SE6
Mg in 5SE5
Mg in 5SE4
Mg in 5SE3
Mg in 5SE2
Mg in 5SE0
Mg in 5SDZ
Mg in 5SE1