Fluorine »
PDB 1bwf-1dvy »
1d4y »
Fluorine in PDB 1d4y: Hiv-1 Protease Triple Mutant/Tipranavir Complex
Enzymatic activity of Hiv-1 Protease Triple Mutant/Tipranavir Complex
All present enzymatic activity of Hiv-1 Protease Triple Mutant/Tipranavir Complex:
3.4.23.16;
3.4.23.16;
Protein crystallography data
The structure of Hiv-1 Protease Triple Mutant/Tipranavir Complex, PDB code: 1d4y
was solved by
K.D.Watenpaugh,
M.N.Janakiraman,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 10.00 / 1.97 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 59.785, 87.503, 46.458, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.4 / 21.6 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Hiv-1 Protease Triple Mutant/Tipranavir Complex
(pdb code 1d4y). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Hiv-1 Protease Triple Mutant/Tipranavir Complex, PDB code: 1d4y:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Hiv-1 Protease Triple Mutant/Tipranavir Complex, PDB code: 1d4y:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 1d4y
Go back to
Fluorine binding site 1 out
of 3 in the Hiv-1 Protease Triple Mutant/Tipranavir Complex
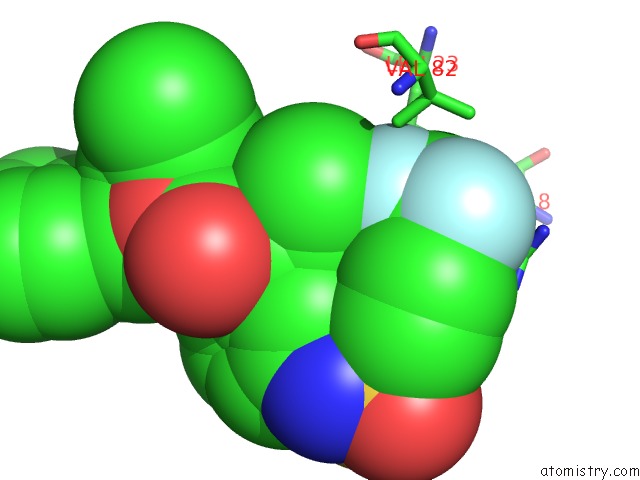
Mono view
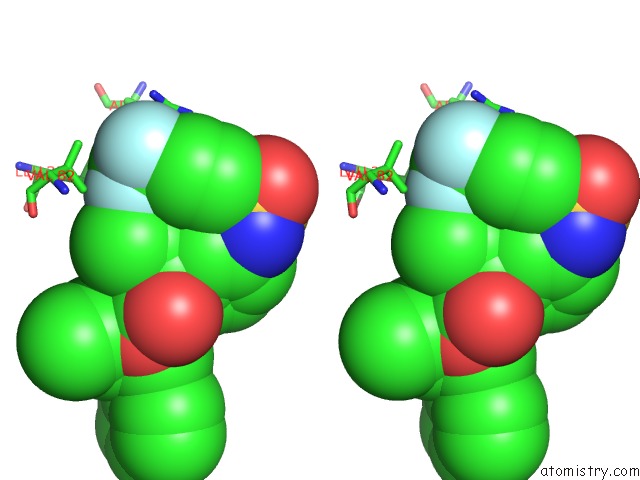
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Hiv-1 Protease Triple Mutant/Tipranavir Complex within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 1d4y
Go back to
Fluorine binding site 2 out
of 3 in the Hiv-1 Protease Triple Mutant/Tipranavir Complex
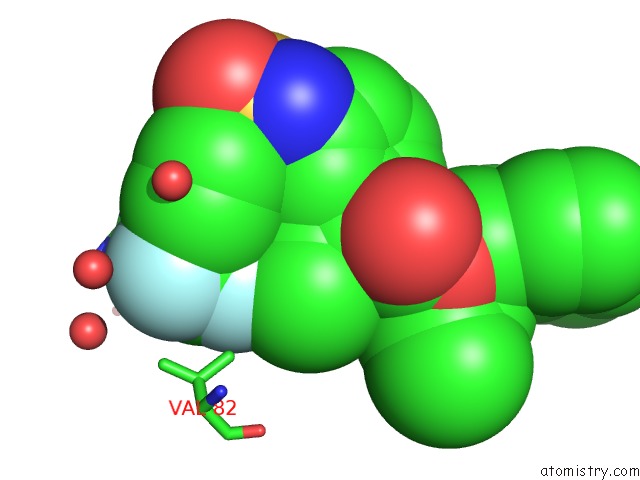
Mono view
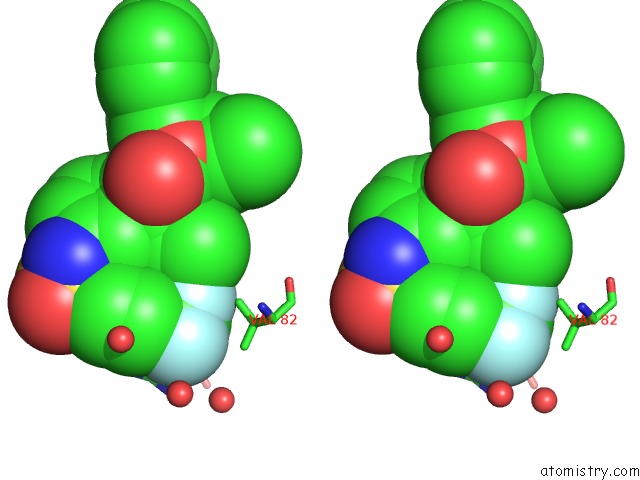
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Hiv-1 Protease Triple Mutant/Tipranavir Complex within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 1d4y
Go back to
Fluorine binding site 3 out
of 3 in the Hiv-1 Protease Triple Mutant/Tipranavir Complex
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Hiv-1 Protease Triple Mutant/Tipranavir Complex within 5.0Å range:
|
Reference:
S.Thaisrivongs,
H.I.Skulnick,
S.R.Turner,
J.W.Strohbach,
R.A.Tommasi,
P.D.Johnson,
P.A.Aristoff,
T.M.Judge,
R.B.Gammill,
J.K.Morris,
K.R.Romines,
R.A.Chrusciel,
R.R.Hinshaw,
K.T.Chong,
W.G.Tarpley,
S.M.Poppe,
D.E.Slade,
J.C.Lynn,
M.M.Horng,
P.K.Tomich,
E.P.Seest,
L.A.Dolak,
W.J.Howe,
G.M.Howard,
K.D.Watenpaugh.
Structure-Based Design of Hiv Protease Inhibitors: Sulfonamide-Containing 5,6-Dihydro-4-Hydroxy-2-Pyrones As Non-Peptidic Inhibitors. J.Med.Chem. V. 39 4349 1996.
ISSN: ISSN 0022-2623
PubMed: 8893827
DOI: 10.1021/JM960541S
Page generated: Wed Jul 31 11:04:19 2024
ISSN: ISSN 0022-2623
PubMed: 8893827
DOI: 10.1021/JM960541S
Last articles
Cl in 5SEDCl in 5SE3
Cl in 5SDS
Cl in 5SEE
Cl in 5SDR
Cl in 5SDQ
Cl in 5SDP
Cl in 5SDN
Cl in 5SDO
Cl in 5SDM