Fluorine »
PDB 1dvz-1fk9 »
1dxw »
Fluorine in PDB 1dxw: Structure of Hetero Complex of Non Structural Protein (Ns) of Hepatitis C Virus (Hcv) and Synthetic Peptidic Compound
Other elements in 1dxw:
The structure of Structure of Hetero Complex of Non Structural Protein (Ns) of Hepatitis C Virus (Hcv) and Synthetic Peptidic Compound also contains other interesting chemical elements:
| Zinc | (Zn) | 20 atoms |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Structure of Hetero Complex of Non Structural Protein (Ns) of Hepatitis C Virus (Hcv) and Synthetic Peptidic Compound
(pdb code 1dxw). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Structure of Hetero Complex of Non Structural Protein (Ns) of Hepatitis C Virus (Hcv) and Synthetic Peptidic Compound, PDB code: 1dxw:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Structure of Hetero Complex of Non Structural Protein (Ns) of Hepatitis C Virus (Hcv) and Synthetic Peptidic Compound, PDB code: 1dxw:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 1dxw
Go back to
Fluorine binding site 1 out
of 2 in the Structure of Hetero Complex of Non Structural Protein (Ns) of Hepatitis C Virus (Hcv) and Synthetic Peptidic Compound
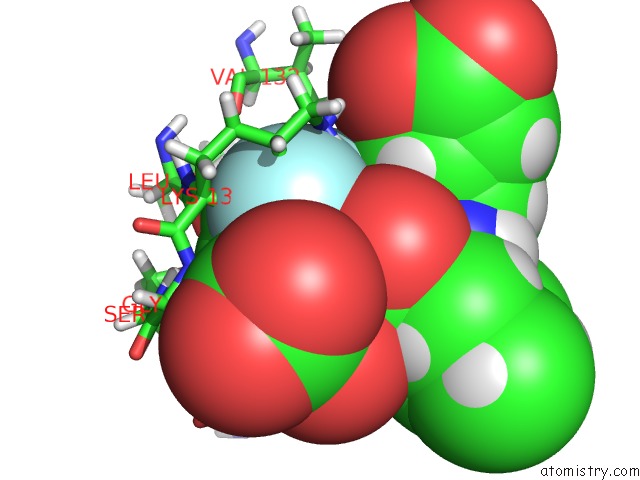
Mono view
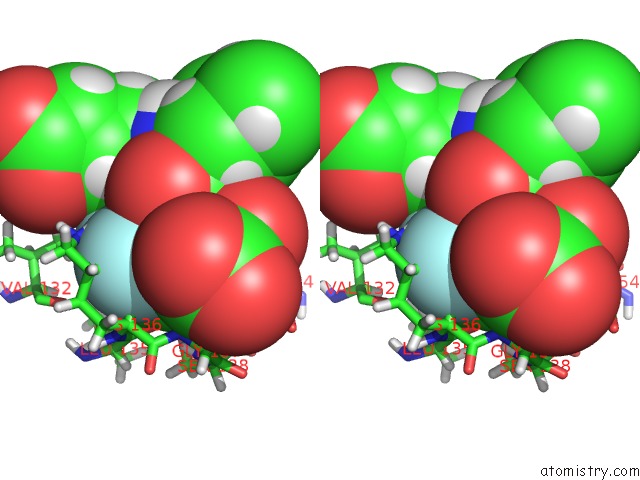
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Structure of Hetero Complex of Non Structural Protein (Ns) of Hepatitis C Virus (Hcv) and Synthetic Peptidic Compound within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 1dxw
Go back to
Fluorine binding site 2 out
of 2 in the Structure of Hetero Complex of Non Structural Protein (Ns) of Hepatitis C Virus (Hcv) and Synthetic Peptidic Compound
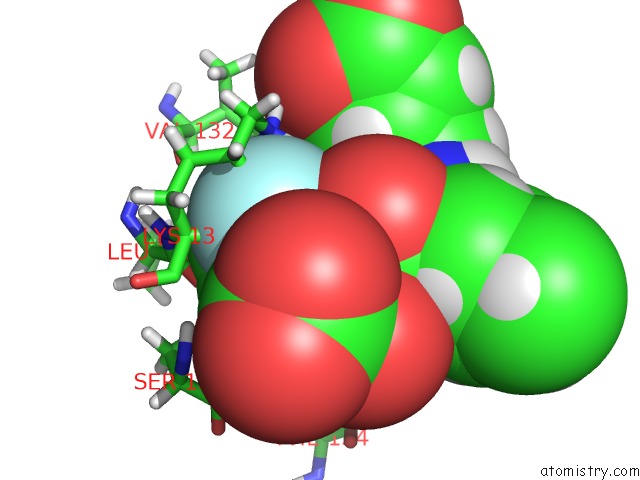
Mono view
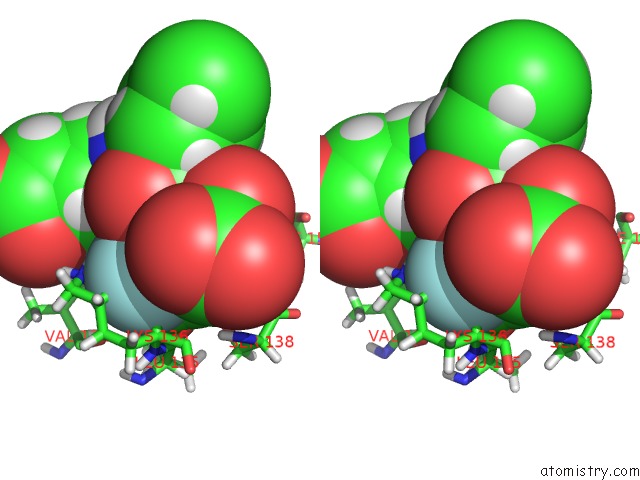
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Structure of Hetero Complex of Non Structural Protein (Ns) of Hepatitis C Virus (Hcv) and Synthetic Peptidic Compound within 5.0Å range:
|
Reference:
G.Barbato,
D.O.Cicero,
F.Cordier,
F.Narjes,
B.Gerlach,
S.Sambucini,
S.Grzesiek,
V.G.Matassa,
R.Defrancesco,
R.Bazzo.
Inhibitor Binding Induces Active Site Stabilisation of the Hcv NS3 Protein Serine Protease Domain Embo J. V. 19 1195 2000.
ISSN: ISSN 0261-4189
PubMed: 10716920
DOI: 10.1093/EMBOJ/19.6.1195
Page generated: Mon Jul 14 10:34:31 2025
ISSN: ISSN 0261-4189
PubMed: 10716920
DOI: 10.1093/EMBOJ/19.6.1195
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO