Fluorine »
PDB 1dvz-1fk9 »
1exx »
Fluorine in PDB 1exx: Enantiomer Discrimination Illustrated By Crystal Structures of the Human Retinoic Acid Receptor Hrargamma Ligand Binding Domain: the Complex with the Inactive S-Enantiomer BMS270395.
Protein crystallography data
The structure of Enantiomer Discrimination Illustrated By Crystal Structures of the Human Retinoic Acid Receptor Hrargamma Ligand Binding Domain: the Complex with the Inactive S-Enantiomer BMS270395., PDB code: 1exx
was solved by
B.P.Klaholz,
A.Mitschler,
M.Belema,
C.Zusi,
D.Moras,
Structural Proteomicsin Europe (Spine),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 6.00 / 1.67 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 59.849, 59.849, 155.615, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.1 / 24.3 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Enantiomer Discrimination Illustrated By Crystal Structures of the Human Retinoic Acid Receptor Hrargamma Ligand Binding Domain: the Complex with the Inactive S-Enantiomer BMS270395.
(pdb code 1exx). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Enantiomer Discrimination Illustrated By Crystal Structures of the Human Retinoic Acid Receptor Hrargamma Ligand Binding Domain: the Complex with the Inactive S-Enantiomer BMS270395., PDB code: 1exx:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Enantiomer Discrimination Illustrated By Crystal Structures of the Human Retinoic Acid Receptor Hrargamma Ligand Binding Domain: the Complex with the Inactive S-Enantiomer BMS270395., PDB code: 1exx:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 1exx
Go back to
Fluorine binding site 1 out
of 2 in the Enantiomer Discrimination Illustrated By Crystal Structures of the Human Retinoic Acid Receptor Hrargamma Ligand Binding Domain: the Complex with the Inactive S-Enantiomer BMS270395.
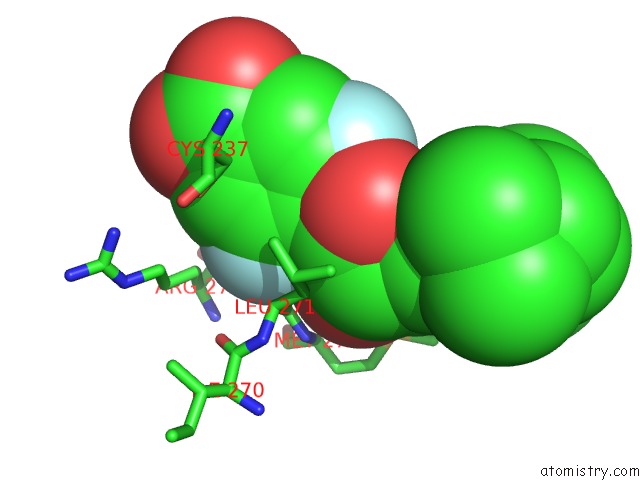
Mono view
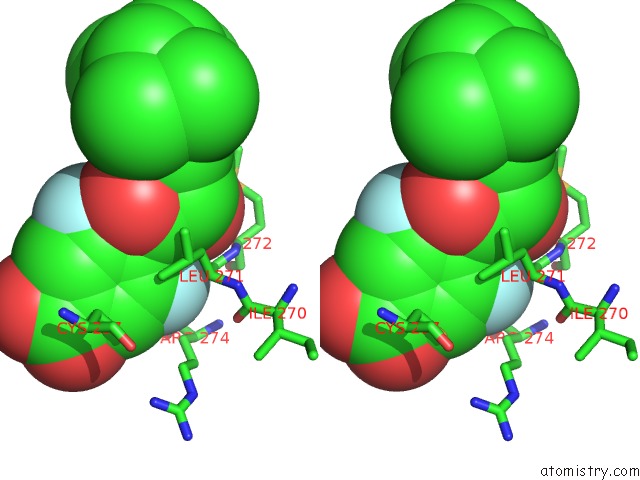
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Enantiomer Discrimination Illustrated By Crystal Structures of the Human Retinoic Acid Receptor Hrargamma Ligand Binding Domain: the Complex with the Inactive S-Enantiomer BMS270395. within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 1exx
Go back to
Fluorine binding site 2 out
of 2 in the Enantiomer Discrimination Illustrated By Crystal Structures of the Human Retinoic Acid Receptor Hrargamma Ligand Binding Domain: the Complex with the Inactive S-Enantiomer BMS270395.
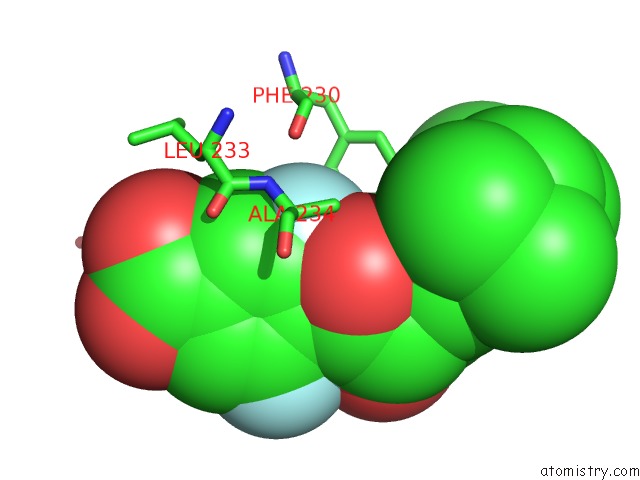
Mono view
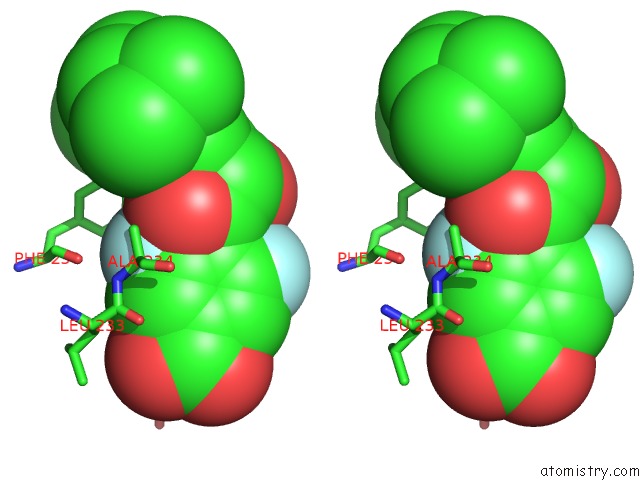
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Enantiomer Discrimination Illustrated By Crystal Structures of the Human Retinoic Acid Receptor Hrargamma Ligand Binding Domain: the Complex with the Inactive S-Enantiomer BMS270395. within 5.0Å range:
|
Reference:
B.P.Klaholz,
A.Mitschler,
M.Belema,
C.Zusi,
D.Moras.
Enantiomer Discrimination Illustrated By High-Resolution Crystal Structures of the Human Nuclear Receptor Hrargamma. Proc.Natl.Acad.Sci.Usa V. 97 6322 2000.
ISSN: ISSN 0027-8424
PubMed: 10841540
DOI: 10.1073/PNAS.97.12.6322
Page generated: Wed Jul 31 11:15:09 2024
ISSN: ISSN 0027-8424
PubMed: 10841540
DOI: 10.1073/PNAS.97.12.6322
Last articles
As in 7SXPAs in 7RP2
As in 7S6U
As in 7RVI
As in 7OI1
As in 7R3F
As in 7QUG
As in 7MDP
As in 7NSE
As in 7NRP