Fluorine »
PDB 1mmd-1o5f »
1mue »
Fluorine in PDB 1mue: Thrombin-Hirugen-L405,426
Enzymatic activity of Thrombin-Hirugen-L405,426
All present enzymatic activity of Thrombin-Hirugen-L405,426:
3.4.21.5;
3.4.21.5;
Protein crystallography data
The structure of Thrombin-Hirugen-L405,426, PDB code: 1mue
was solved by
C.S.Burgey,
K.A.Robinson,
T.A.Lyle,
P.G.Nantermet,
H.G.Selnick,
R.C.Isaacs,
S.D.Lewis,
B.J.Lucas,
J.A.Krueger,
R.Singh,
C.Miller-Stein,
R.B.White,
B.Wong,
E.A.Lyle,
M.T.Stranieri,
J.J.Cook,
D.R.Mcmasters,
J.M.Pellicore,
S.Pal,
A.A.Wallace,
F.C.Clayton,
D.Bohn,
D.C.Welsh,
J.J.Lynch,
Y.Yan,
Z.Chen,
L.Kuo,
S.J.Gardell,
J.A.Shafer,
J.P.Vacca,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 2.00 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 71.010, 72.110, 72.840, 90.00, 100.66, 90.00 |
| R / Rfree (%) | 20.1 / n/a |
Other elements in 1mue:
The structure of Thrombin-Hirugen-L405,426 also contains other interesting chemical elements:
| Chlorine | (Cl) | 1 atom |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Thrombin-Hirugen-L405,426
(pdb code 1mue). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Thrombin-Hirugen-L405,426, PDB code: 1mue:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Thrombin-Hirugen-L405,426, PDB code: 1mue:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 1mue
Go back to
Fluorine binding site 1 out
of 3 in the Thrombin-Hirugen-L405,426
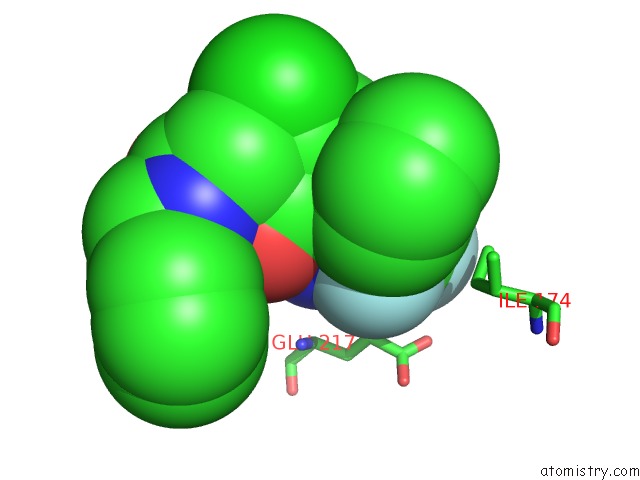
Mono view
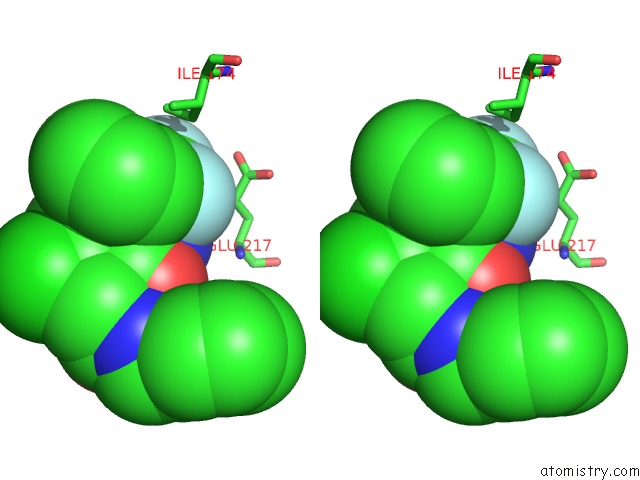
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Thrombin-Hirugen-L405,426 within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 1mue
Go back to
Fluorine binding site 2 out
of 3 in the Thrombin-Hirugen-L405,426
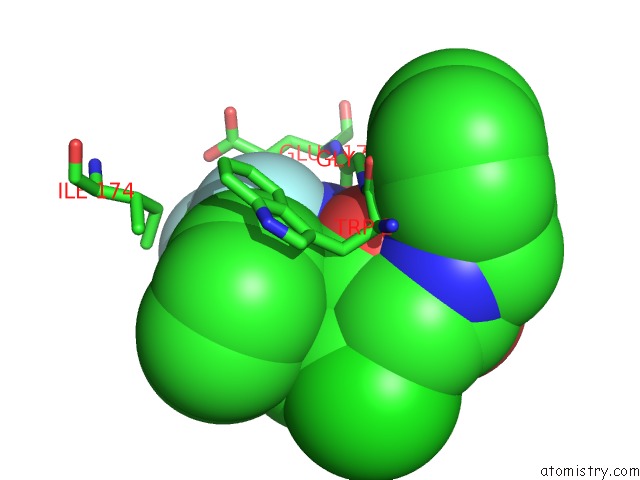
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Thrombin-Hirugen-L405,426 within 5.0Å range:
|
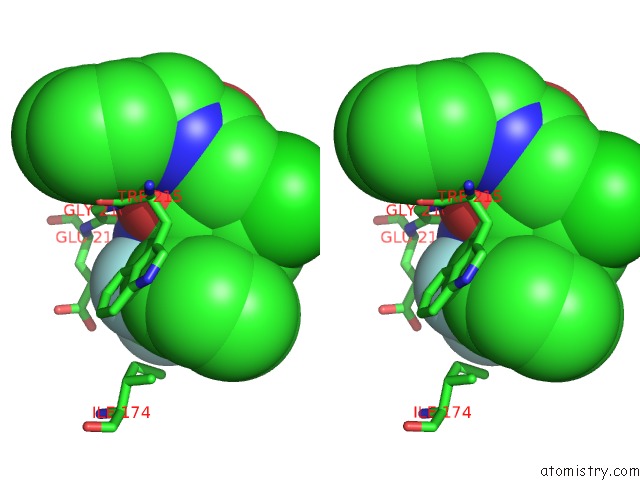
Fluorine binding site 3 out of 3 in 1mue
Go back to
Fluorine binding site 3 out
of 3 in the Thrombin-Hirugen-L405,426
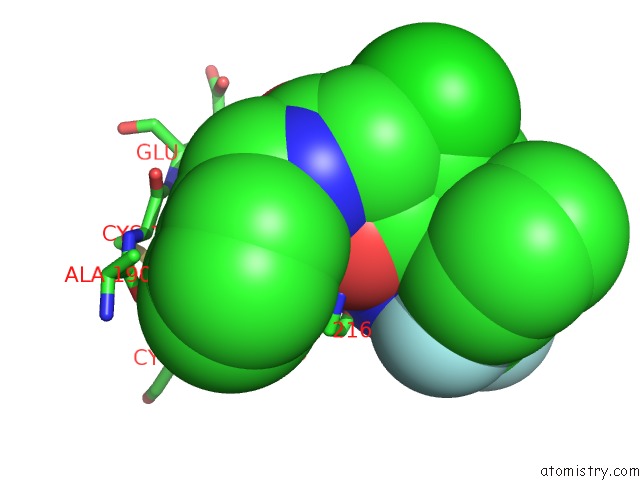
Mono view
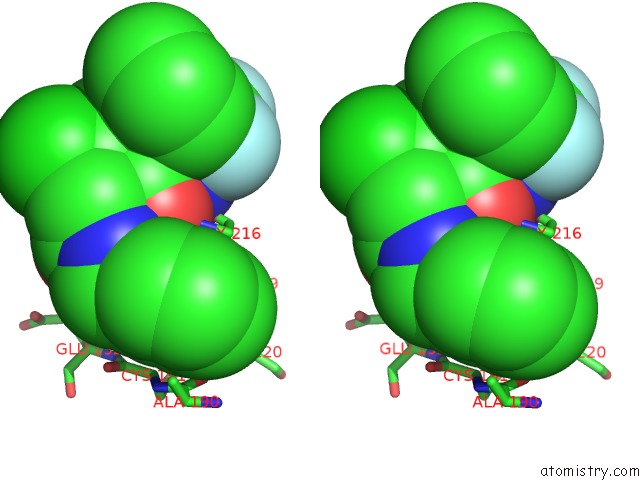
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Thrombin-Hirugen-L405,426 within 5.0Å range:
|
Reference:
C.S.Burgey,
K.A.Robinson,
T.A.Lyle,
P.G.Nantermet,
H.G.Selnick,
R.C.Isaacs,
S.D.Lewis,
B.J.Lucas,
J.A.Krueger,
R.Singh,
C.Miller-Stein,
R.B.White,
B.Wong,
E.A.Lyle,
M.T.Stranieri,
J.J.Cook,
D.R.Mcmasters,
J.M.Pellicore,
S.Pal,
A.A.Wallace,
F.C.Clayton,
D.Bohn,
D.C.Welsh,
J.J.Lynch,
Y.Yan,
Z.Chen,
L.Kuo,
S.J.Gardell,
J.A.Shafer,
J.P.Vacca.
Pharmacokinetic Optimization of 3-Amino-6-Chloropyrazinone Acetamide Thrombin Inhibitors. Implementation of P3 Pyridine N-Oxides to Deliver An Orally Bioavailable Series Containing P1 N-Benzylamides. Bioorg.Med.Chem.Lett. V. 13 1353 2003.
ISSN: ISSN 0960-894X
PubMed: 12657281
DOI: 10.1016/S0960-894X(03)00099-4
Page generated: Mon Jul 14 11:19:44 2025
ISSN: ISSN 0960-894X
PubMed: 12657281
DOI: 10.1016/S0960-894X(03)00099-4
Last articles
F in 3ECCF in 3E7T
F in 3E7C
F in 3E2M
F in 3E7I
F in 3E6T
F in 3E6O
F in 3DZF
F in 3E65
F in 3DZQ