Fluorine »
PDB 1mmd-1o5f »
1n0q »
Fluorine in PDB 1n0q: 3ANK: A Designed Ankyrin Repeat Protein with Three Identical Consensus Repeats
Protein crystallography data
The structure of 3ANK: A Designed Ankyrin Repeat Protein with Three Identical Consensus Repeats, PDB code: 1n0q
was solved by
L.K.Mosavi,
D.L.Minor Jr.,
Z.-Y.Peng,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.96 / 1.26 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 39.636, 43.159, 105.574, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.1 / 18.7 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the 3ANK: A Designed Ankyrin Repeat Protein with Three Identical Consensus Repeats
(pdb code 1n0q). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the 3ANK: A Designed Ankyrin Repeat Protein with Three Identical Consensus Repeats, PDB code: 1n0q:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the 3ANK: A Designed Ankyrin Repeat Protein with Three Identical Consensus Repeats, PDB code: 1n0q:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 1n0q
Go back to
Fluorine binding site 1 out
of 3 in the 3ANK: A Designed Ankyrin Repeat Protein with Three Identical Consensus Repeats
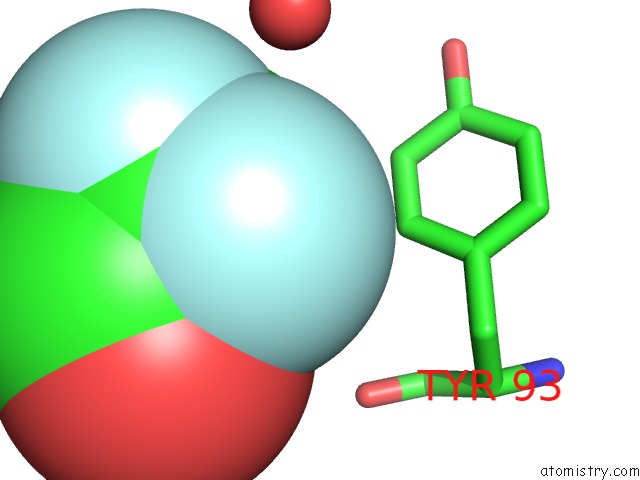
Mono view
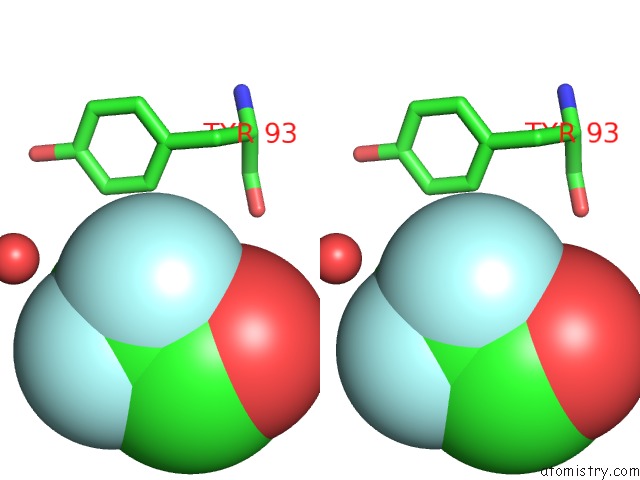
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of 3ANK: A Designed Ankyrin Repeat Protein with Three Identical Consensus Repeats within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 1n0q
Go back to
Fluorine binding site 2 out
of 3 in the 3ANK: A Designed Ankyrin Repeat Protein with Three Identical Consensus Repeats
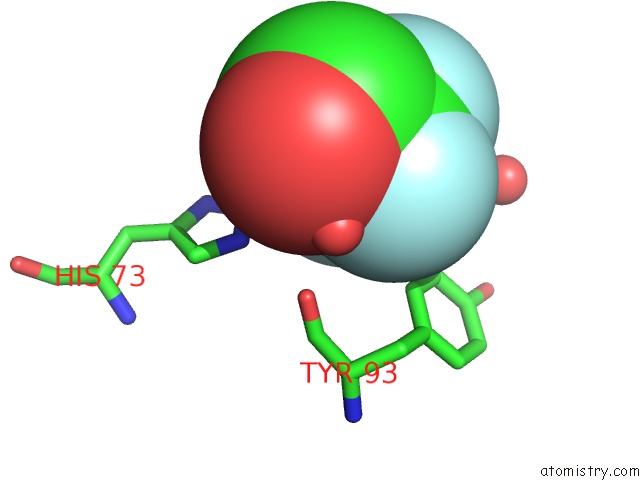
Mono view
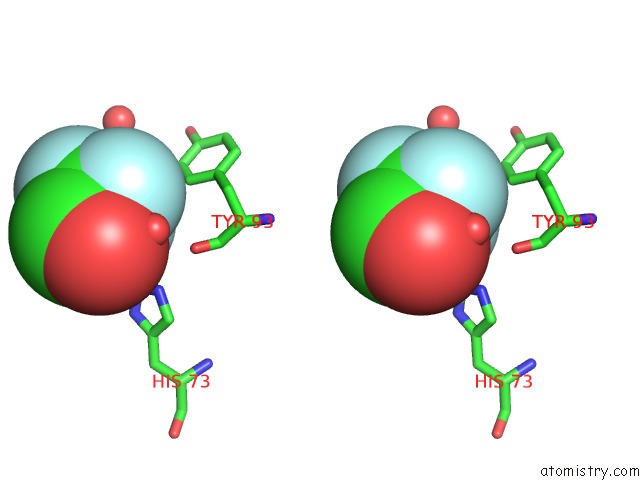
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of 3ANK: A Designed Ankyrin Repeat Protein with Three Identical Consensus Repeats within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 1n0q
Go back to
Fluorine binding site 3 out
of 3 in the 3ANK: A Designed Ankyrin Repeat Protein with Three Identical Consensus Repeats
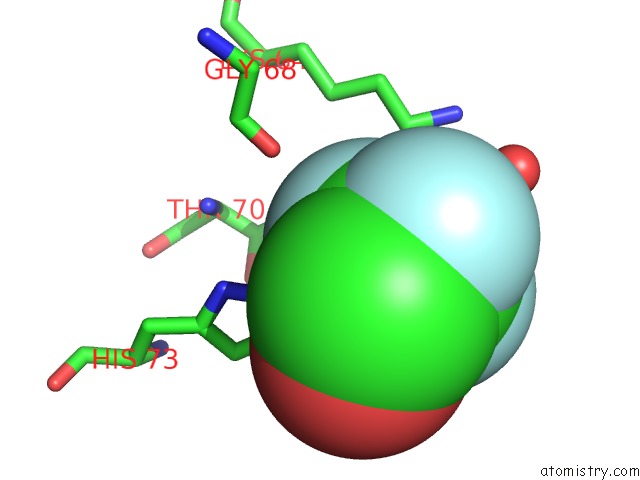
Mono view
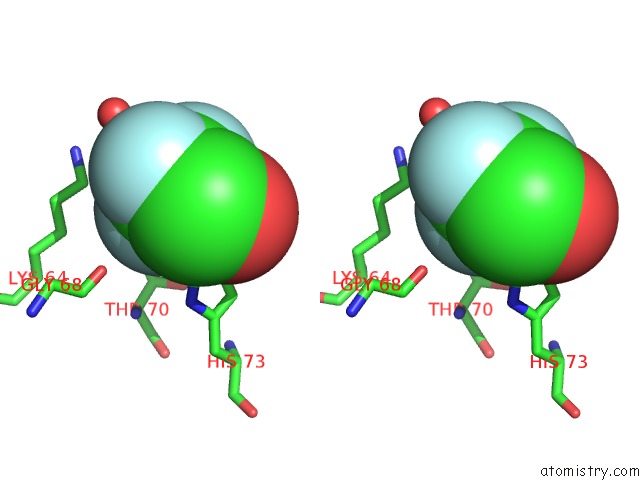
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of 3ANK: A Designed Ankyrin Repeat Protein with Three Identical Consensus Repeats within 5.0Å range:
|
Reference:
L.K.Mosavi,
D.L.Minor Jr.,
Z.-Y.Peng.
Consensus-Derived Structural Determinants of the Ankyrin Repeat Motif. Proc.Natl.Acad.Sci.Usa V. 99 16029 2002.
ISSN: ISSN 0027-8424
PubMed: 12461176
DOI: 10.1073/PNAS.252537899
Page generated: Wed Jul 31 12:05:09 2024
ISSN: ISSN 0027-8424
PubMed: 12461176
DOI: 10.1073/PNAS.252537899
Last articles
Ca in 5UN2Ca in 5UU7
Ca in 5UQZ
Ca in 5ULY
Ca in 5UP7
Ca in 5UN3
Ca in 5UO9
Ca in 5ULU
Ca in 5UKG
Ca in 5UMH