Fluorine »
PDB 1udb-1w5y »
1uwj »
Fluorine in PDB 1uwj: The Complex of Mutant V599E B-Raf and BAY439006.
Enzymatic activity of The Complex of Mutant V599E B-Raf and BAY439006.
All present enzymatic activity of The Complex of Mutant V599E B-Raf and BAY439006.:
2.7.1.37; 2.7.11.1;
2.7.1.37; 2.7.11.1;
Protein crystallography data
The structure of The Complex of Mutant V599E B-Raf and BAY439006., PDB code: 1uwj
was solved by
D.Barford,
S.M.Roe,
P.T.C.Wan,
Cancer Genome Project,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.39 / 3.50 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 97.660, 97.660, 168.070, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 27.5 / 35.7 |
Other elements in 1uwj:
The structure of The Complex of Mutant V599E B-Raf and BAY439006. also contains other interesting chemical elements:
| Chlorine | (Cl) | 2 atoms |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the The Complex of Mutant V599E B-Raf and BAY439006.
(pdb code 1uwj). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 6 binding sites of Fluorine where determined in the The Complex of Mutant V599E B-Raf and BAY439006., PDB code: 1uwj:
Jump to Fluorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Fluorine where determined in the The Complex of Mutant V599E B-Raf and BAY439006., PDB code: 1uwj:
Jump to Fluorine binding site number: 1; 2; 3; 4; 5; 6;
Fluorine binding site 1 out of 6 in 1uwj
Go back to
Fluorine binding site 1 out
of 6 in the The Complex of Mutant V599E B-Raf and BAY439006.
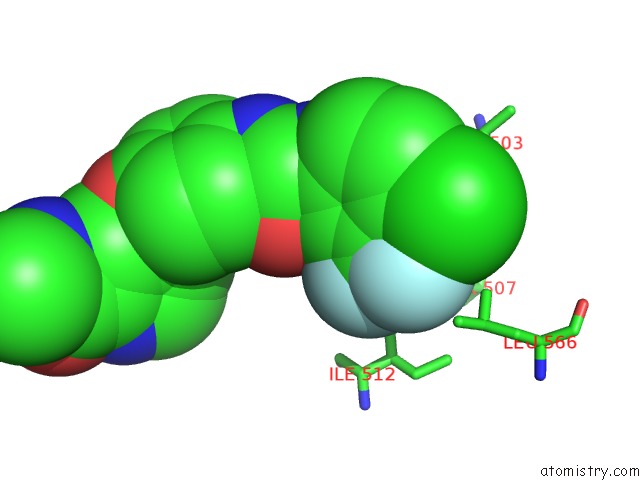
Mono view
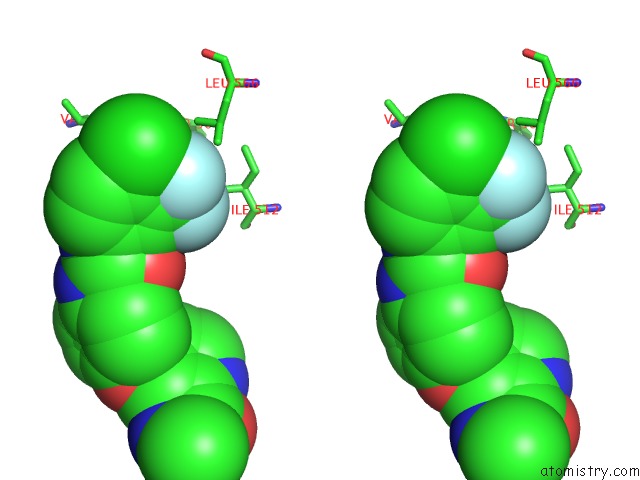
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of The Complex of Mutant V599E B-Raf and BAY439006. within 5.0Å range:
|
Fluorine binding site 2 out of 6 in 1uwj
Go back to
Fluorine binding site 2 out
of 6 in the The Complex of Mutant V599E B-Raf and BAY439006.
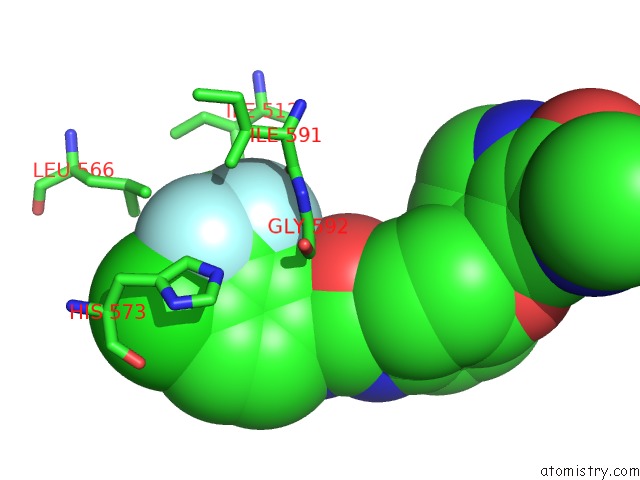
Mono view
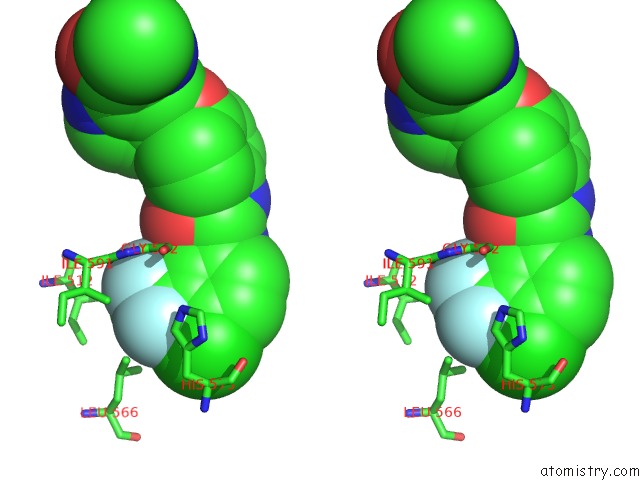
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of The Complex of Mutant V599E B-Raf and BAY439006. within 5.0Å range:
|
Fluorine binding site 3 out of 6 in 1uwj
Go back to
Fluorine binding site 3 out
of 6 in the The Complex of Mutant V599E B-Raf and BAY439006.
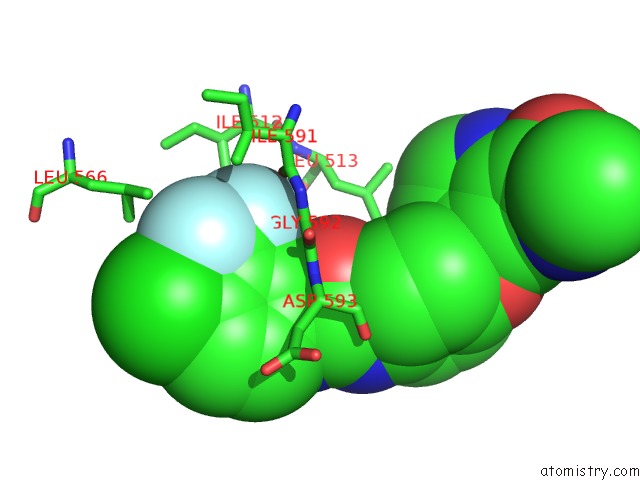
Mono view
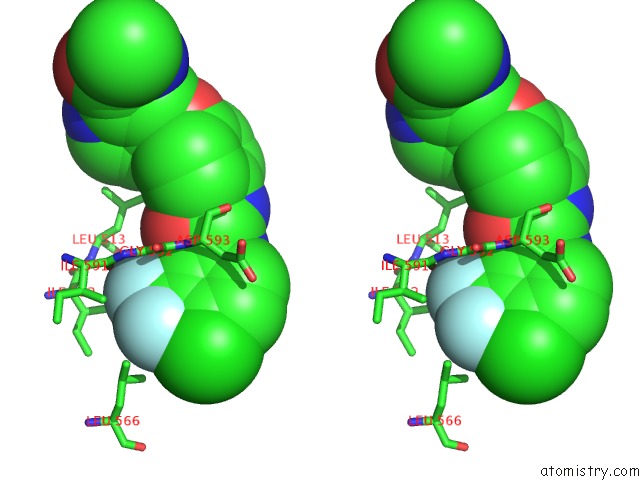
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of The Complex of Mutant V599E B-Raf and BAY439006. within 5.0Å range:
|
Fluorine binding site 4 out of 6 in 1uwj
Go back to
Fluorine binding site 4 out
of 6 in the The Complex of Mutant V599E B-Raf and BAY439006.
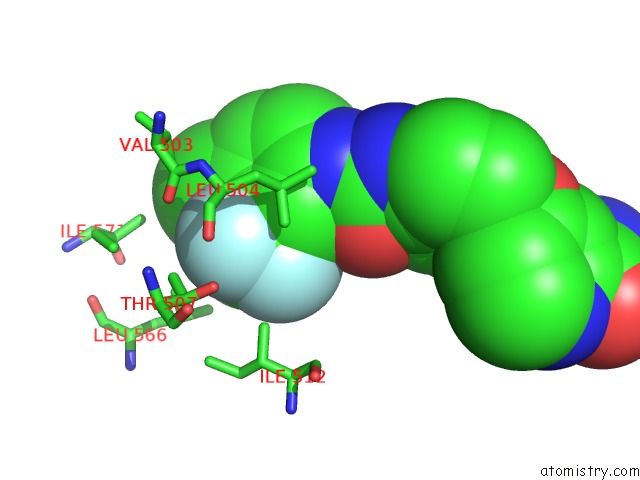
Mono view
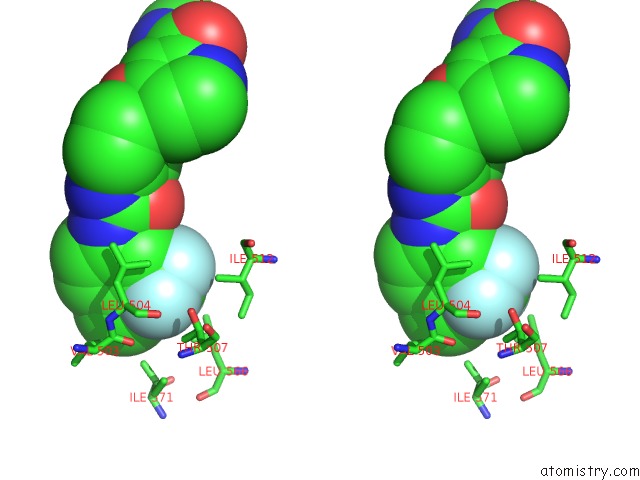
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 4 of The Complex of Mutant V599E B-Raf and BAY439006. within 5.0Å range:
|
Fluorine binding site 5 out of 6 in 1uwj
Go back to
Fluorine binding site 5 out
of 6 in the The Complex of Mutant V599E B-Raf and BAY439006.
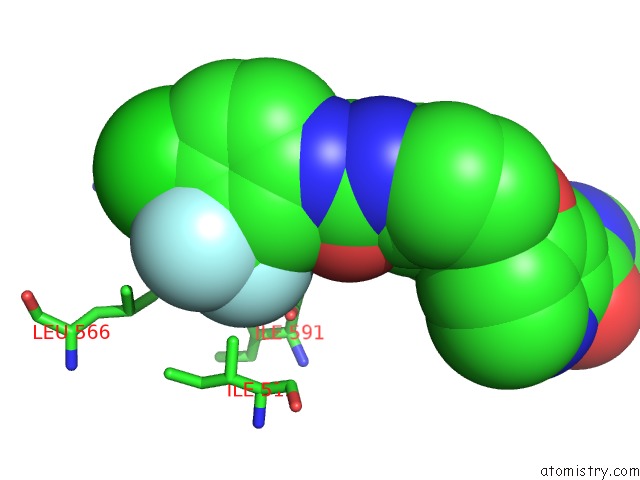
Mono view
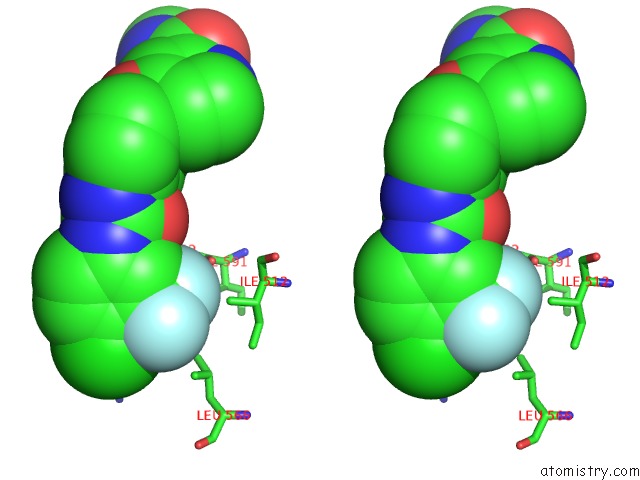
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 5 of The Complex of Mutant V599E B-Raf and BAY439006. within 5.0Å range:
|
Fluorine binding site 6 out of 6 in 1uwj
Go back to
Fluorine binding site 6 out
of 6 in the The Complex of Mutant V599E B-Raf and BAY439006.
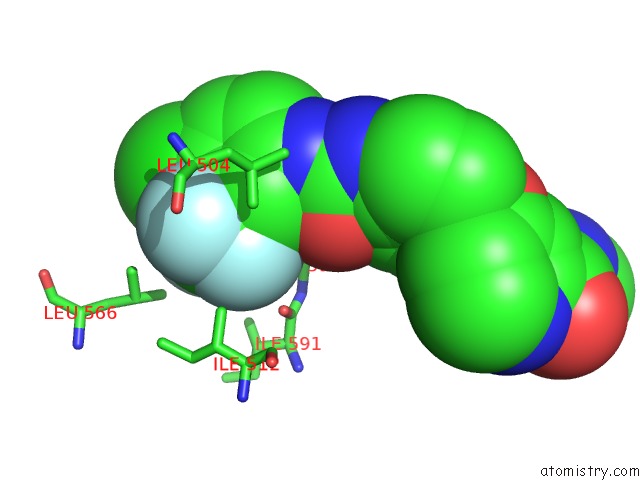
Mono view
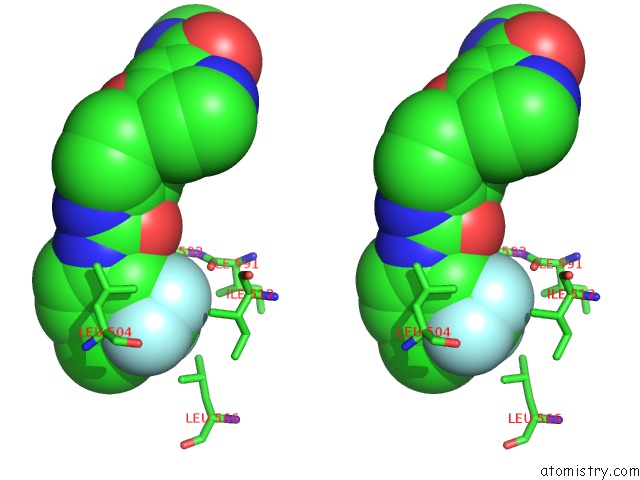
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 6 of The Complex of Mutant V599E B-Raf and BAY439006. within 5.0Å range:
|
Reference:
P.T.C.Wan,
M.J.Garnett,
S.M.Roe,
S.Lee,
D.Niculescu-Duvaz,
V.M.Good,
C.M.Jones,
C.J.Marshall,
C.J.Springer,
D.Barford,
R.Marais.
Mechanism of Activation of the Raf-Erk Signaling Pathway By Oncogenic Mutations of B-Raf Cell(Cambridge,Mass.) V. 116 855 2004.
ISSN: ISSN 0092-8674
PubMed: 15035987
DOI: 10.1016/S0092-8674(04)00215-6
Page generated: Mon Jul 14 12:05:49 2025
ISSN: ISSN 0092-8674
PubMed: 15035987
DOI: 10.1016/S0092-8674(04)00215-6
Last articles
F in 7K0WF in 7JYR
F in 7JY3
F in 7JYQ
F in 7JX9
F in 7JY1
F in 7JY0
F in 7JXW
F in 7JXZ
F in 7JXK