Fluorine »
PDB 2oh4-2pdk »
2pdi »
Fluorine in PDB 2pdi: Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A.
Enzymatic activity of Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A.
All present enzymatic activity of Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A.:
1.1.1.21;
1.1.1.21;
Protein crystallography data
The structure of Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A., PDB code: 2pdi
was solved by
H.Steuber,
A.Heine,
G.Klebe,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.55 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.440, 66.860, 47.300, 90.00, 92.95, 90.00 |
| R / Rfree (%) | 15.5 / 20 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A.
(pdb code 2pdi). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A., PDB code: 2pdi:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A., PDB code: 2pdi:
Jump to Fluorine binding site number: 1; 2; 3;
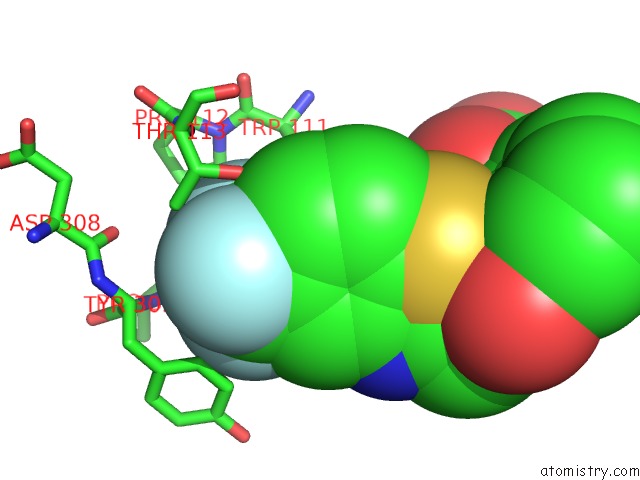
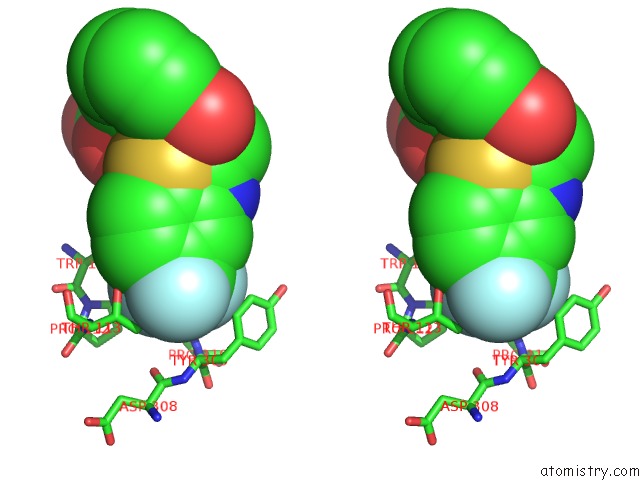
Fluorine binding site 1 out of 3 in 2pdi
Go back to
Fluorine binding site 1 out
of 3 in the Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A. within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 2pdi
Go back to
Fluorine binding site 2 out
of 3 in the Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A.

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A. within 5.0Å range:
|
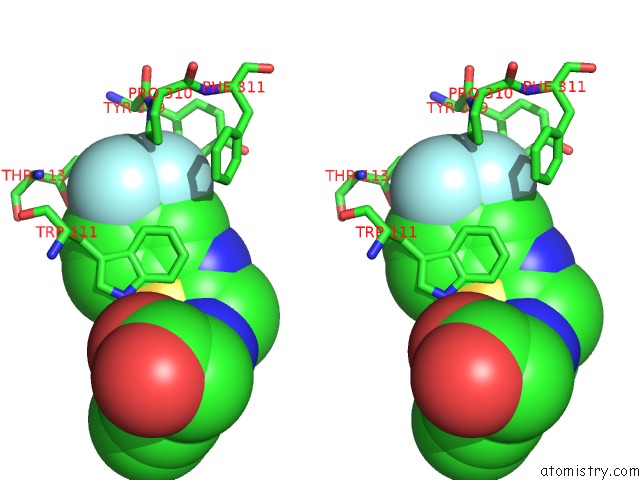
Fluorine binding site 3 out of 3 in 2pdi
Go back to
Fluorine binding site 3 out
of 3 in the Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A.

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Human Aldose Reductase Mutant L300A Complexed with Zopolrestat at 1.55 A. within 5.0Å range:
|
Reference:
H.Steuber,
A.Heine,
A.Podjarny,
G.Klebe.
Merging the Binding Sites of Aldose and Aldehyde Reductase For Detection of Inhibitor Selectivity-Determining Features. J.Mol.Biol. V. 379 991 2008.
ISSN: ISSN 0022-2836
PubMed: 18495158
DOI: 10.1016/J.JMB.2008.03.063
Page generated: Mon Jul 14 13:58:11 2025
ISSN: ISSN 0022-2836
PubMed: 18495158
DOI: 10.1016/J.JMB.2008.03.063
Last articles
Fe in 2B11Fe in 2B12
Fe in 2B10
Fe in 2AYS
Fe in 2B0Z
Fe in 2AZQ
Fe in 2AXT
Fe in 2AV8
Fe in 2AWY
Fe in 2AXX