Fluorine »
PDB 3ccw-3d39 »
3ce3 »
Fluorine in PDB 3ce3: Crystal Structure of the Tyrosine Kinase Domain of the Hepatocyte Growth Factor Receptor C-Met in Complex with A Pyrrolopyridinepyridone Based Inhibitor
Enzymatic activity of Crystal Structure of the Tyrosine Kinase Domain of the Hepatocyte Growth Factor Receptor C-Met in Complex with A Pyrrolopyridinepyridone Based Inhibitor
All present enzymatic activity of Crystal Structure of the Tyrosine Kinase Domain of the Hepatocyte Growth Factor Receptor C-Met in Complex with A Pyrrolopyridinepyridone Based Inhibitor:
2.7.10.1;
2.7.10.1;
Protein crystallography data
The structure of Crystal Structure of the Tyrosine Kinase Domain of the Hepatocyte Growth Factor Receptor C-Met in Complex with A Pyrrolopyridinepyridone Based Inhibitor, PDB code: 3ce3
was solved by
J.Sack,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.40 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 43.040, 49.137, 159.324, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.6 / 27.5 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of the Tyrosine Kinase Domain of the Hepatocyte Growth Factor Receptor C-Met in Complex with A Pyrrolopyridinepyridone Based Inhibitor
(pdb code 3ce3). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Crystal Structure of the Tyrosine Kinase Domain of the Hepatocyte Growth Factor Receptor C-Met in Complex with A Pyrrolopyridinepyridone Based Inhibitor, PDB code: 3ce3:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Crystal Structure of the Tyrosine Kinase Domain of the Hepatocyte Growth Factor Receptor C-Met in Complex with A Pyrrolopyridinepyridone Based Inhibitor, PDB code: 3ce3:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 3ce3
Go back to
Fluorine binding site 1 out
of 2 in the Crystal Structure of the Tyrosine Kinase Domain of the Hepatocyte Growth Factor Receptor C-Met in Complex with A Pyrrolopyridinepyridone Based Inhibitor
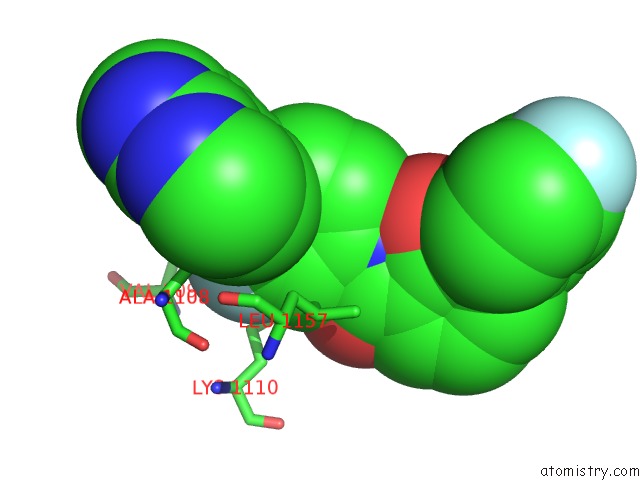
Mono view
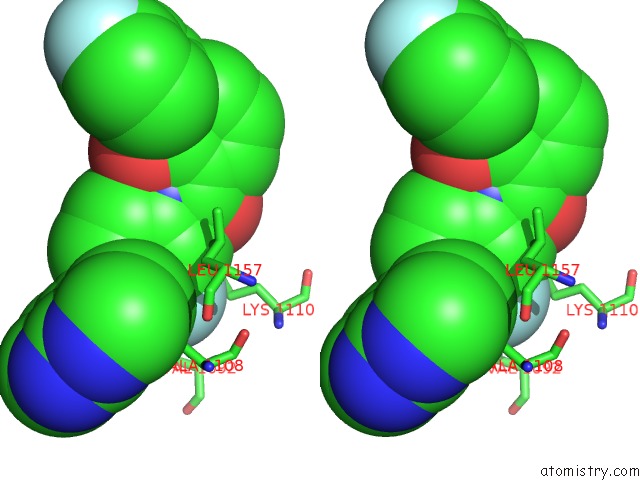
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of the Tyrosine Kinase Domain of the Hepatocyte Growth Factor Receptor C-Met in Complex with A Pyrrolopyridinepyridone Based Inhibitor within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 3ce3
Go back to
Fluorine binding site 2 out
of 2 in the Crystal Structure of the Tyrosine Kinase Domain of the Hepatocyte Growth Factor Receptor C-Met in Complex with A Pyrrolopyridinepyridone Based Inhibitor
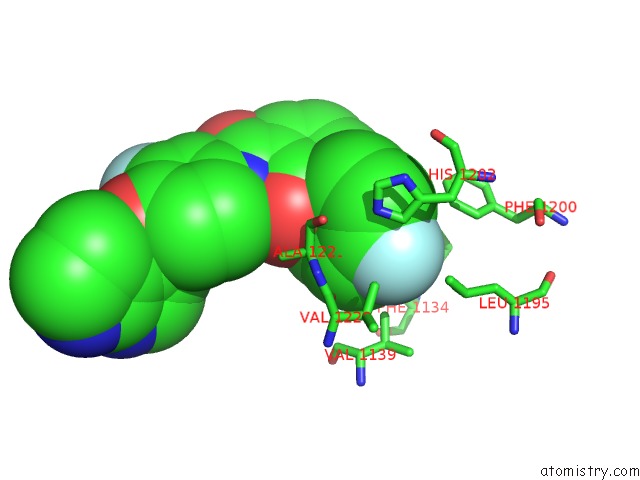
Mono view
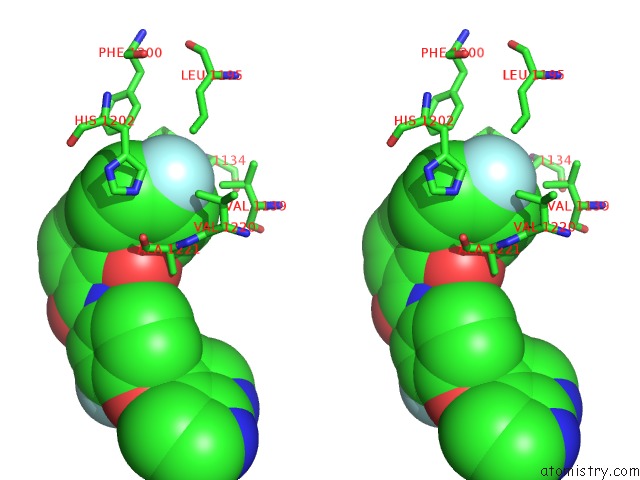
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of the Tyrosine Kinase Domain of the Hepatocyte Growth Factor Receptor C-Met in Complex with A Pyrrolopyridinepyridone Based Inhibitor within 5.0Å range:
|
Reference:
K.S.Kim,
L.Zhang,
R.Schmidt,
Z.W.Cai,
D.Wei,
D.K.Williams,
L.J.Lombardo,
G.L.Trainor,
D.Xie,
Y.Zhang,
Y.An,
J.S.Sack,
J.S.Tokarski,
C.Darienzo,
A.Kamath,
P.Marathe,
Y.Zhang,
J.Lippy,
R.Jeyaseelan,
B.Wautlet,
B.Henley,
J.Gullo-Brown,
V.Manne,
J.T.Hunt,
J.Fargnoli,
R.M.Borzilleri.
Discovery of Pyrrolopyridine-Pyridone Based Inhibitors of Met Kinase: Synthesis, X-Ray Crystallographic Analysis, and Biological Activities. J.Med.Chem. V. 51 5330 2008.
ISSN: ISSN 0022-2623
PubMed: 18690676
DOI: 10.1021/JM800476Q
Page generated: Mon Jul 14 15:27:22 2025
ISSN: ISSN 0022-2623
PubMed: 18690676
DOI: 10.1021/JM800476Q
Last articles
F in 7KHLF in 7KHH
F in 7KHG
F in 7KH6
F in 7KBG
F in 7KBN
F in 7KBC
F in 7KAF
F in 7KA1
F in 7K77