Fluorine »
PDB 3d3e-3du8 »
3d5m »
Fluorine in PDB 3d5m: Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor
Enzymatic activity of Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor
All present enzymatic activity of Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor:
2.7.7.48;
2.7.7.48;
Protein crystallography data
The structure of Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor, PDB code: 3d5m
was solved by
Q.Zhao,
R.E.Showalter,
Q.Han,
C.R.Kissinger,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.00 / 2.20 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 86.304, 106.765, 126.254, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.8 / 26.1 |
Other elements in 3d5m:
The structure of Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor also contains other interesting chemical elements:
| Chlorine | (Cl) | 2 atoms |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor
(pdb code 3d5m). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor, PDB code: 3d5m:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor, PDB code: 3d5m:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 3d5m
Go back to
Fluorine binding site 1 out
of 2 in the Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor
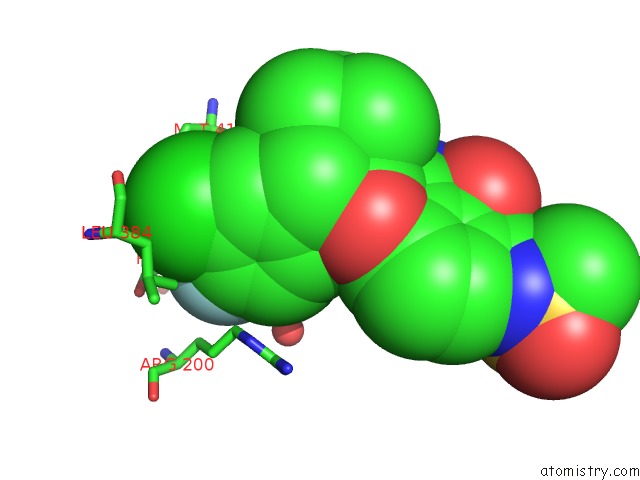
Mono view
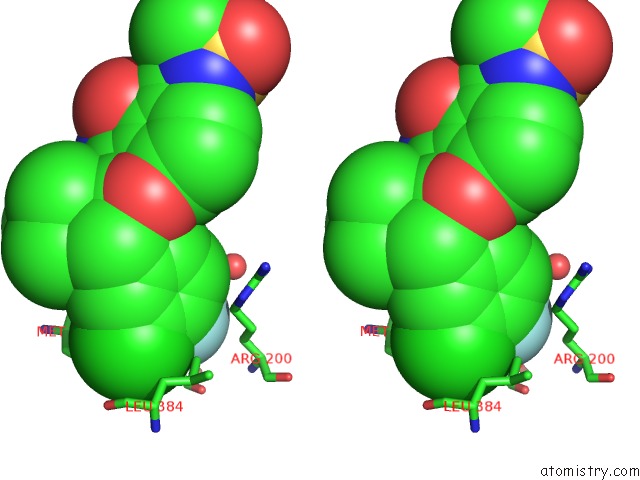
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 3d5m
Go back to
Fluorine binding site 2 out
of 2 in the Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor
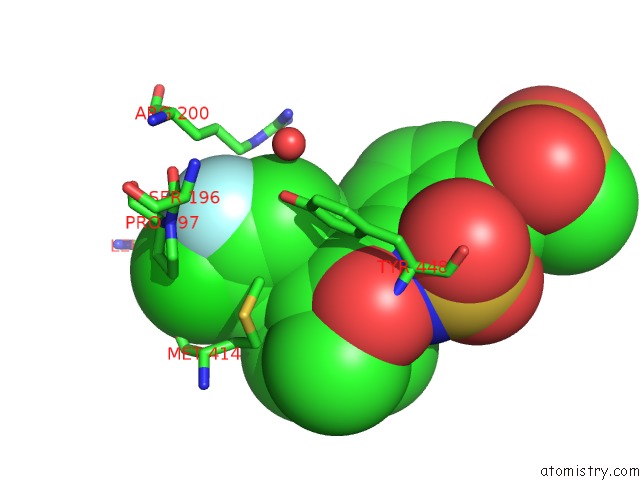
Mono view
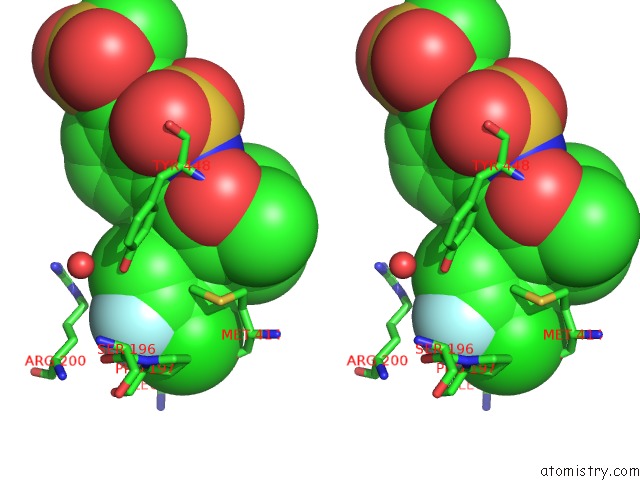
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of Hcv NS5B Polymerase with A Novel Pyridazinone Inhibitor within 5.0Å range:
|
Reference:
S.H.Kim,
M.T.Tran,
F.Ruebsam,
A.X.Xiang,
B.Ayida,
H.Mcguire,
D.Ellis,
J.Blazel,
C.V.Tran,
D.E.Murphy,
S.E.Webber,
Y.Zhou,
A.M.Shah,
M.Tsan,
R.E.Showalter,
R.Patel,
A.Gobbi,
L.A.Lebrun,
D.M.Bartkowski,
T.G.Nolan,
D.A.Norris,
M.V.Sergeeva,
L.Kirkovsky,
Q.Zhao,
Q.Han,
C.R.Kissinger.
Structure-Based Design, Synthesis, and Biological Evaluation of 1,1-Dioxoisothiazole and Benzo[B]Thiophene-1,1-Dioxide Derivatives As Novel Inhibitors of Hepatitis C Virus NS5B Polymerase. Bioorg.Med.Chem.Lett. V. 18 4181 2008.
ISSN: ISSN 0960-894X
PubMed: 18554907
DOI: 10.1016/J.BMCL.2008.05.083
Page generated: Mon Jul 14 15:46:28 2025
ISSN: ISSN 0960-894X
PubMed: 18554907
DOI: 10.1016/J.BMCL.2008.05.083
Last articles
F in 7LCRF in 7LCM
F in 7LCO
F in 7LCK
F in 7LCJ
F in 7LCI
F in 7L9Y
F in 7LCD
F in 7LAY
F in 7LAJ