Fluorine »
PDB 3g72-3gwv »
3gnr »
Fluorine in PDB 3gnr: Crystal Structure of A Rice OS3BGLU6 Beta-Glucosidase with Covalently Bound 2-Deoxy-2-Fluoroglucoside to the Catalytic Nucleophile E396
Enzymatic activity of Crystal Structure of A Rice OS3BGLU6 Beta-Glucosidase with Covalently Bound 2-Deoxy-2-Fluoroglucoside to the Catalytic Nucleophile E396
All present enzymatic activity of Crystal Structure of A Rice OS3BGLU6 Beta-Glucosidase with Covalently Bound 2-Deoxy-2-Fluoroglucoside to the Catalytic Nucleophile E396:
3.2.1.21;
3.2.1.21;
Protein crystallography data
The structure of Crystal Structure of A Rice OS3BGLU6 Beta-Glucosidase with Covalently Bound 2-Deoxy-2-Fluoroglucoside to the Catalytic Nucleophile E396, PDB code: 3gnr
was solved by
S.Seshadri,
T.Akiyama,
R.Opassiri,
B.Kuaprasert,
J.R.K.Cairns,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 27.74 / 1.81 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 56.679, 90.591, 101.883, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.2 / 18.5 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of A Rice OS3BGLU6 Beta-Glucosidase with Covalently Bound 2-Deoxy-2-Fluoroglucoside to the Catalytic Nucleophile E396
(pdb code 3gnr). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total only one binding site of Fluorine was determined in the Crystal Structure of A Rice OS3BGLU6 Beta-Glucosidase with Covalently Bound 2-Deoxy-2-Fluoroglucoside to the Catalytic Nucleophile E396, PDB code: 3gnr:
In total only one binding site of Fluorine was determined in the Crystal Structure of A Rice OS3BGLU6 Beta-Glucosidase with Covalently Bound 2-Deoxy-2-Fluoroglucoside to the Catalytic Nucleophile E396, PDB code: 3gnr:
Fluorine binding site 1 out of 1 in 3gnr
Go back to
Fluorine binding site 1 out
of 1 in the Crystal Structure of A Rice OS3BGLU6 Beta-Glucosidase with Covalently Bound 2-Deoxy-2-Fluoroglucoside to the Catalytic Nucleophile E396
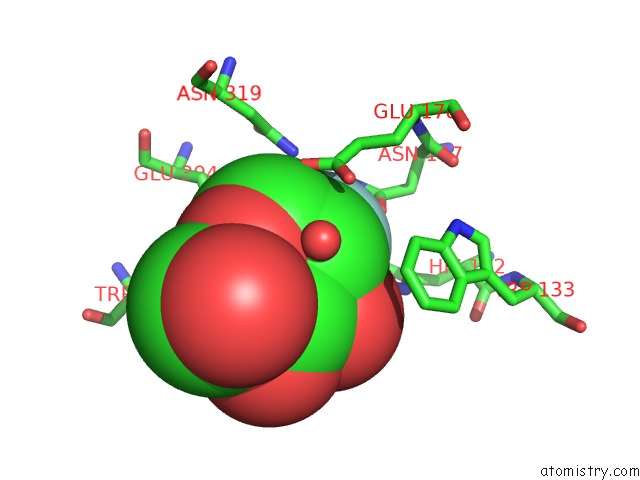
Mono view
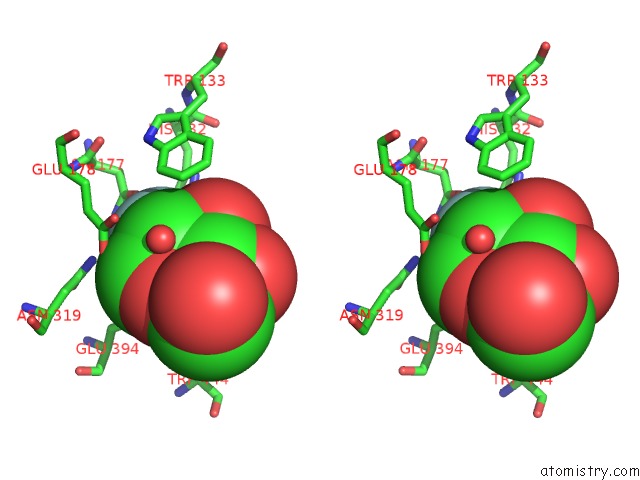
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of A Rice OS3BGLU6 Beta-Glucosidase with Covalently Bound 2-Deoxy-2-Fluoroglucoside to the Catalytic Nucleophile E396 within 5.0Å range:
|
Reference:
S.Seshadri,
T.Akiyama,
R.Opassiri,
B.Kuaprasert,
J.R.K.Cairns.
Structural and Enzymatic Characterization of OS3BGLU6, A Rice Beta-Glucosidase Hydrolyzing Hydrophobic Glycosides and (1->3)- and (1->2)-Linked Disaccharides. Plant Physiol. V. 151 47 2009.
ISSN: ISSN 0032-0889
PubMed: 19587102
DOI: 10.1104/PP.109.139436
Page generated: Wed Jul 31 18:56:12 2024
ISSN: ISSN 0032-0889
PubMed: 19587102
DOI: 10.1104/PP.109.139436
Last articles
Cl in 7TJOCl in 7TLE
Cl in 7TKV
Cl in 7THH
Cl in 7TIV
Cl in 7TIW
Cl in 7TI9
Cl in 7TIU
Cl in 7TIN
Cl in 7TI7