Fluorine »
PDB 3jx3-3kql »
3kou »
Fluorine in PDB 3kou: Structural Insights Into the Catalytic Mechanism of CD38: Evidence For A Conformationally Flexible Covalent Enzyme-Substrate Complex.
Enzymatic activity of Structural Insights Into the Catalytic Mechanism of CD38: Evidence For A Conformationally Flexible Covalent Enzyme-Substrate Complex.
All present enzymatic activity of Structural Insights Into the Catalytic Mechanism of CD38: Evidence For A Conformationally Flexible Covalent Enzyme-Substrate Complex.:
3.2.2.5;
3.2.2.5;
Protein crystallography data
The structure of Structural Insights Into the Catalytic Mechanism of CD38: Evidence For A Conformationally Flexible Covalent Enzyme-Substrate Complex., PDB code: 3kou
was solved by
P.F.Egea,
H.Muller-Steffner,
R.M.Stroud,
N.J.Oppenheimer,
E.Kellenberger,
F.Schuber,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.64 / 1.78 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 46.721, 81.287, 150.909, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20 / 24.3 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Structural Insights Into the Catalytic Mechanism of CD38: Evidence For A Conformationally Flexible Covalent Enzyme-Substrate Complex.
(pdb code 3kou). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Structural Insights Into the Catalytic Mechanism of CD38: Evidence For A Conformationally Flexible Covalent Enzyme-Substrate Complex., PDB code: 3kou:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Structural Insights Into the Catalytic Mechanism of CD38: Evidence For A Conformationally Flexible Covalent Enzyme-Substrate Complex., PDB code: 3kou:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 3kou
Go back to
Fluorine binding site 1 out
of 2 in the Structural Insights Into the Catalytic Mechanism of CD38: Evidence For A Conformationally Flexible Covalent Enzyme-Substrate Complex.
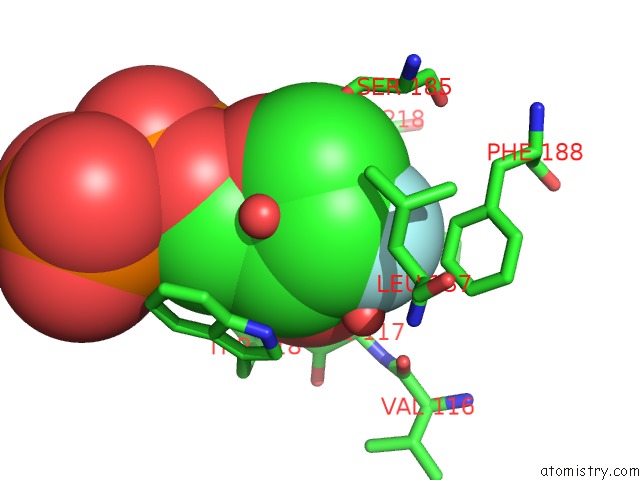
Mono view
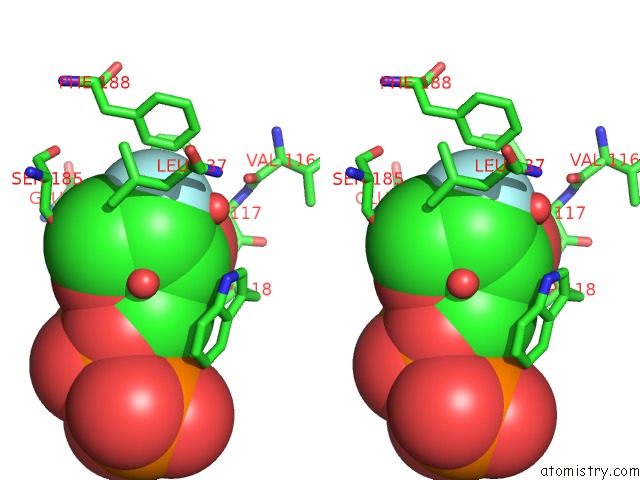
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Structural Insights Into the Catalytic Mechanism of CD38: Evidence For A Conformationally Flexible Covalent Enzyme-Substrate Complex. within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 3kou
Go back to
Fluorine binding site 2 out
of 2 in the Structural Insights Into the Catalytic Mechanism of CD38: Evidence For A Conformationally Flexible Covalent Enzyme-Substrate Complex.
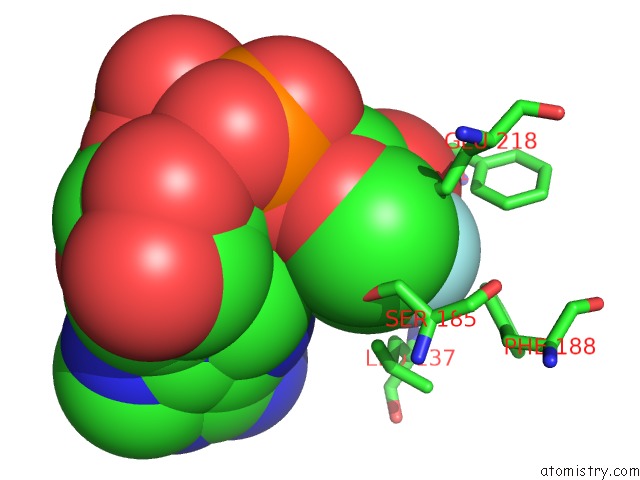
Mono view
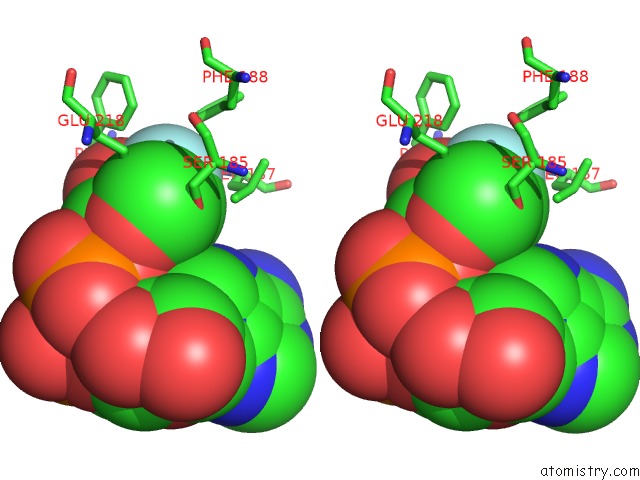
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Structural Insights Into the Catalytic Mechanism of CD38: Evidence For A Conformationally Flexible Covalent Enzyme-Substrate Complex. within 5.0Å range:
|
Reference:
P.F.Egea,
H.Muller-Steffner,
I.Kuhn,
C.Cakir-Kiefer,
N.J.Oppenheimer,
R.M.Stroud,
E.Kellenberger,
F.Schuber.
Insights Into the Mechanism of Bovine CD38/Nad+Glycohydrolase From the X-Ray Structures of Its Michaelis Complex and Covalently-Trapped Intermediates. Plos One V. 7 34918 2012.
ISSN: ESSN 1932-6203
PubMed: 22529956
DOI: 10.1371/JOURNAL.PONE.0034918
Page generated: Mon Jul 14 17:20:35 2025
ISSN: ESSN 1932-6203
PubMed: 22529956
DOI: 10.1371/JOURNAL.PONE.0034918
Last articles
F in 4GJ3F in 4GHT
F in 4GJ2
F in 4GHI
F in 4GG7
F in 4GG5
F in 4GE6
F in 4GE5
F in 4GCA
F in 4GE2