Fluorine »
PDB 3qrj-3rgf »
3qsb »
Fluorine in PDB 3qsb: Structure of E. Coli Poliiibeta with (Z)-5-(1-((4'-Fluorobiphenyl-4- Yl)Methoxyimino)Butyl)-2,2-Dimethyl-4,6-Dioxocyclohexanecarbonitrile
Enzymatic activity of Structure of E. Coli Poliiibeta with (Z)-5-(1-((4'-Fluorobiphenyl-4- Yl)Methoxyimino)Butyl)-2,2-Dimethyl-4,6-Dioxocyclohexanecarbonitrile
All present enzymatic activity of Structure of E. Coli Poliiibeta with (Z)-5-(1-((4'-Fluorobiphenyl-4- Yl)Methoxyimino)Butyl)-2,2-Dimethyl-4,6-Dioxocyclohexanecarbonitrile:
2.7.7.7;
2.7.7.7;
Protein crystallography data
The structure of Structure of E. Coli Poliiibeta with (Z)-5-(1-((4'-Fluorobiphenyl-4- Yl)Methoxyimino)Butyl)-2,2-Dimethyl-4,6-Dioxocyclohexanecarbonitrile, PDB code: 3qsb
was solved by
G.Wijffels,
W.M.Johnson,
A.J.Oakley,
K.Turner,
V.C.Epa,
S.J.Briscoe,
M.Polley,
A.J.Liepa,
A.Hofmann,
J.Buchardt,
C.Christensen,
P.Prosselkov,
B.P.Dalrymple,
P.F.Alewood,
P.A.Jennings,
N.E.Dixon,
D.A.Winkler,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.69 / 1.90 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 79.176, 66.519, 83.117, 90.00, 115.86, 90.00 |
| R / Rfree (%) | 24.2 / 31.7 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Structure of E. Coli Poliiibeta with (Z)-5-(1-((4'-Fluorobiphenyl-4- Yl)Methoxyimino)Butyl)-2,2-Dimethyl-4,6-Dioxocyclohexanecarbonitrile
(pdb code 3qsb). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Structure of E. Coli Poliiibeta with (Z)-5-(1-((4'-Fluorobiphenyl-4- Yl)Methoxyimino)Butyl)-2,2-Dimethyl-4,6-Dioxocyclohexanecarbonitrile, PDB code: 3qsb:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Structure of E. Coli Poliiibeta with (Z)-5-(1-((4'-Fluorobiphenyl-4- Yl)Methoxyimino)Butyl)-2,2-Dimethyl-4,6-Dioxocyclohexanecarbonitrile, PDB code: 3qsb:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 3qsb
Go back to
Fluorine binding site 1 out
of 2 in the Structure of E. Coli Poliiibeta with (Z)-5-(1-((4'-Fluorobiphenyl-4- Yl)Methoxyimino)Butyl)-2,2-Dimethyl-4,6-Dioxocyclohexanecarbonitrile
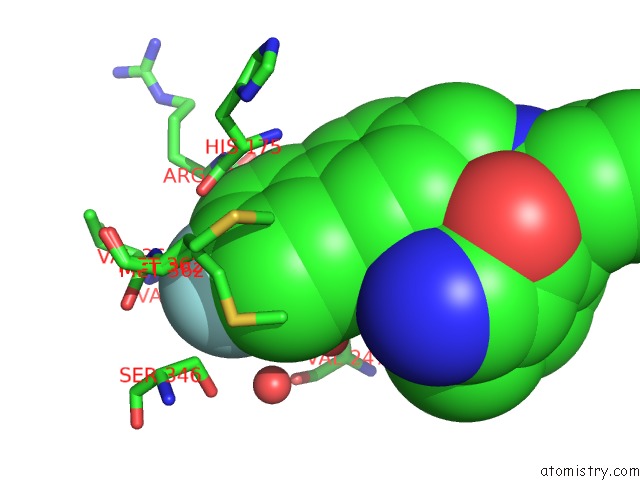
Mono view
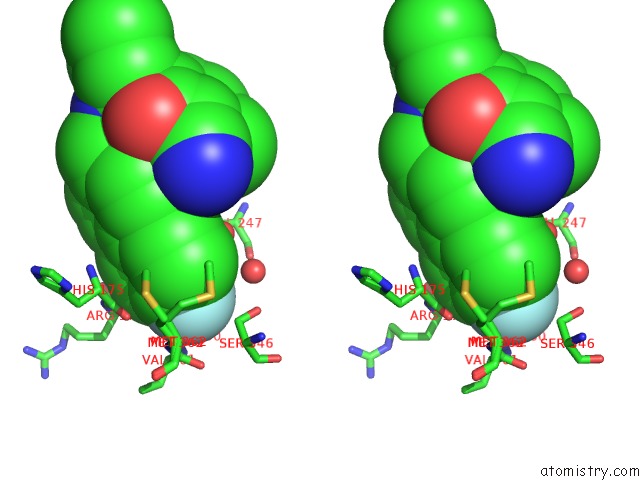
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Structure of E. Coli Poliiibeta with (Z)-5-(1-((4'-Fluorobiphenyl-4- Yl)Methoxyimino)Butyl)-2,2-Dimethyl-4,6-Dioxocyclohexanecarbonitrile within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 3qsb
Go back to
Fluorine binding site 2 out
of 2 in the Structure of E. Coli Poliiibeta with (Z)-5-(1-((4'-Fluorobiphenyl-4- Yl)Methoxyimino)Butyl)-2,2-Dimethyl-4,6-Dioxocyclohexanecarbonitrile
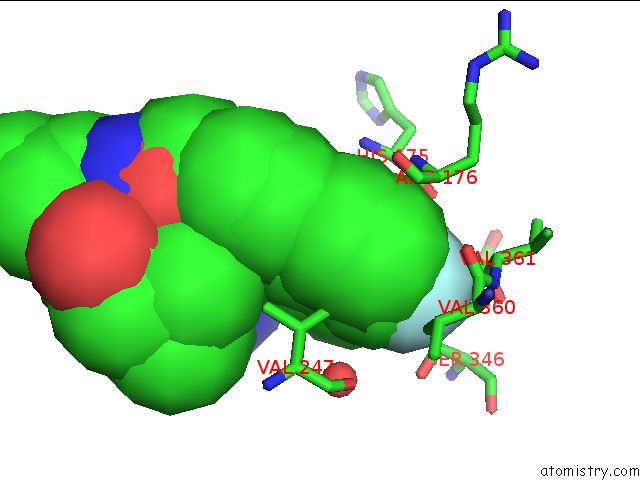
Mono view
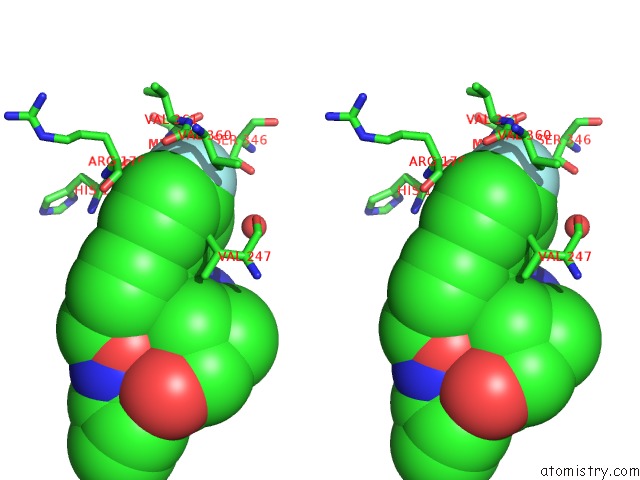
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Structure of E. Coli Poliiibeta with (Z)-5-(1-((4'-Fluorobiphenyl-4- Yl)Methoxyimino)Butyl)-2,2-Dimethyl-4,6-Dioxocyclohexanecarbonitrile within 5.0Å range:
|
Reference:
G.Wijffels,
W.M.Johnson,
A.J.Oakley,
K.Turner,
V.C.Epa,
S.J.Briscoe,
M.Polley,
A.J.Liepa,
A.Hofmann,
J.Buchardt,
C.Christensen,
P.Prosselkov,
B.P.Dalrymple,
P.F.Alewood,
P.A.Jennings,
N.E.Dixon,
D.A.Winkler.
Binding Inhibitors of the Bacterial Sliding Clamp By Design J.Med.Chem. V. 54 4831 2011.
ISSN: ISSN 0022-2623
PubMed: 21604761
DOI: 10.1021/JM2004333
Page generated: Mon Jul 14 19:00:43 2025
ISSN: ISSN 0022-2623
PubMed: 21604761
DOI: 10.1021/JM2004333
Last articles
F in 4HN4F in 4HJX
F in 4HLH
F in 4HL4
F in 4HIQ
F in 4HHZ
F in 4HHY
F in 4HGT
F in 4HEJ
F in 4HGS