Fluorine »
PDB 3qrj-3rgf »
3qt8 »
Fluorine in PDB 3qt8: Crystal Structure of Mutant S192A Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Complexed with Inhibitor 6- Fmvapp
Enzymatic activity of Crystal Structure of Mutant S192A Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Complexed with Inhibitor 6- Fmvapp
All present enzymatic activity of Crystal Structure of Mutant S192A Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Complexed with Inhibitor 6- Fmvapp:
4.1.1.33;
4.1.1.33;
Protein crystallography data
The structure of Crystal Structure of Mutant S192A Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Complexed with Inhibitor 6- Fmvapp, PDB code: 3qt8
was solved by
M.L.Barta,
A.D.Skaff,
W.J.Mcwhorter,
H.M.Miziorko,
B.V.Geisbrecht,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.00 / 2.10 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 82.505, 101.367, 155.255, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19 / 24.1 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of Mutant S192A Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Complexed with Inhibitor 6- Fmvapp
(pdb code 3qt8). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Crystal Structure of Mutant S192A Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Complexed with Inhibitor 6- Fmvapp, PDB code: 3qt8:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Crystal Structure of Mutant S192A Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Complexed with Inhibitor 6- Fmvapp, PDB code: 3qt8:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 3qt8
Go back to
Fluorine binding site 1 out
of 2 in the Crystal Structure of Mutant S192A Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Complexed with Inhibitor 6- Fmvapp
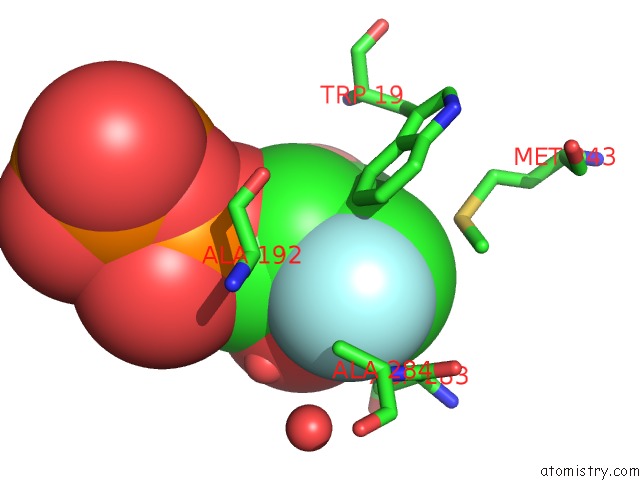
Mono view
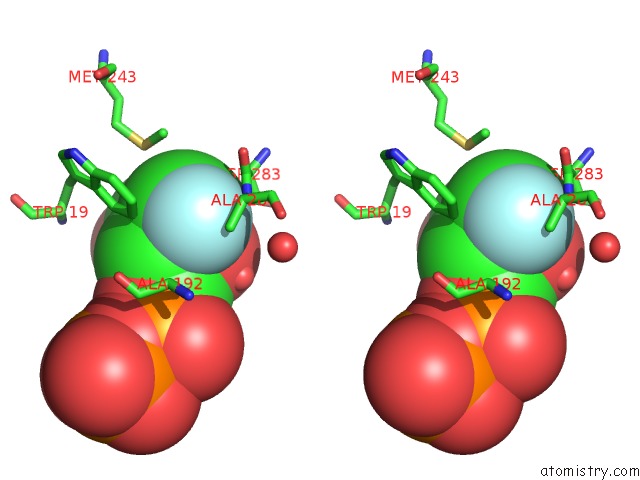
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of Mutant S192A Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Complexed with Inhibitor 6- Fmvapp within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 3qt8
Go back to
Fluorine binding site 2 out
of 2 in the Crystal Structure of Mutant S192A Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Complexed with Inhibitor 6- Fmvapp
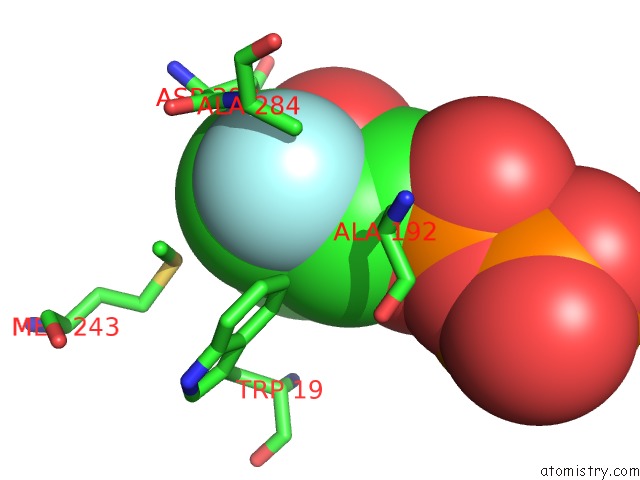
Mono view
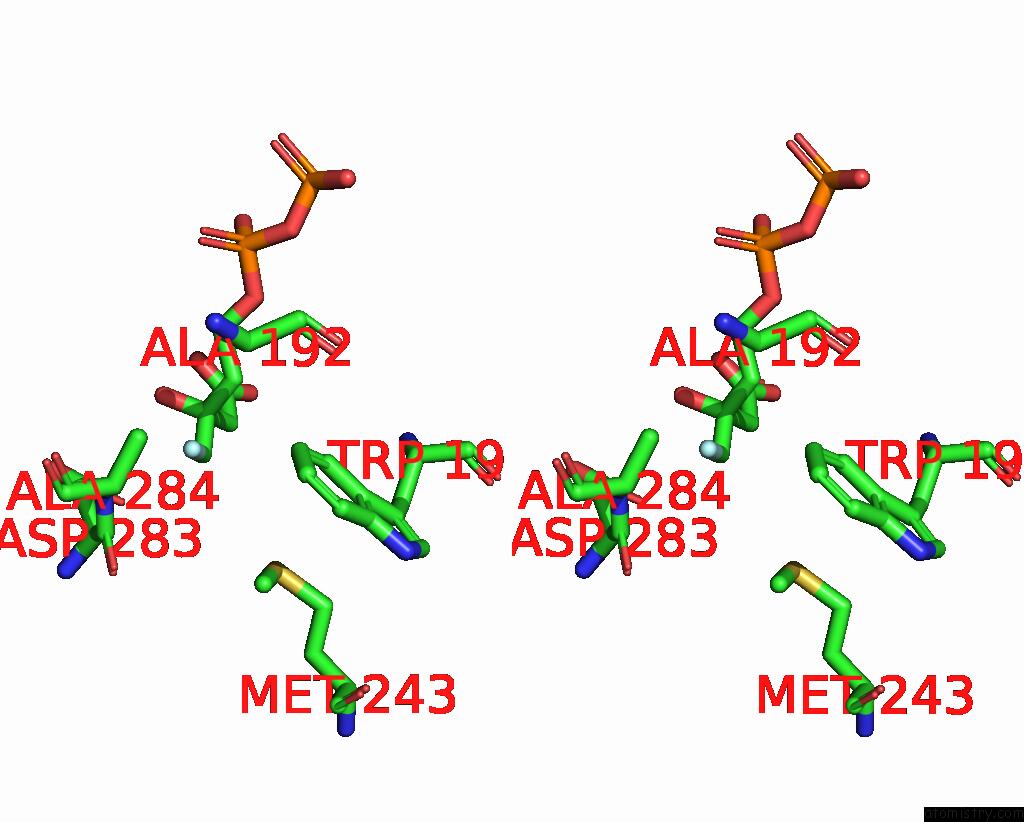
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of Mutant S192A Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Complexed with Inhibitor 6- Fmvapp within 5.0Å range:
|
Reference:
M.L.Barta,
D.A.Skaff,
W.J.Mcwhorter,
T.J.Herdendorf,
H.M.Miziorko,
B.V.Geisbrecht.
Crystal Structures of Staphylococcus Epidermidis Mevalonate Diphosphate Decarboxylase Bound to Inhibitory Analogs Reveal New Insight Into Substrate Binding and Catalysis. J.Biol.Chem. V. 286 23900 2011.
ISSN: ISSN 0021-9258
PubMed: 21561869
DOI: 10.1074/JBC.M111.242016
Page generated: Wed Jul 31 22:08:53 2024
ISSN: ISSN 0021-9258
PubMed: 21561869
DOI: 10.1074/JBC.M111.242016
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW