Fluorine »
PDB 3vrs-3wyk »
3wru »
Fluorine in PDB 3wru: Crystal Structure of the Bacterial Ribosomal Decoding Site in Complex with Synthetic Aminoglycoside with F-Haba Group
Protein crystallography data
The structure of Crystal Structure of the Bacterial Ribosomal Decoding Site in Complex with Synthetic Aminoglycoside with F-Haba Group, PDB code: 3wru
was solved by
J.P.Maianti,
H.Kanazawa,
P.Dozzo,
L.A.Feeney,
E.S.Armstrong,
J.Kondo,
S.Hanessian,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.70 / 2.30 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 46.554, 32.602, 51.413, 90.00, 108.50, 90.00 |
| R / Rfree (%) | 24.9 / 26.2 |
Other elements in 3wru:
The structure of Crystal Structure of the Bacterial Ribosomal Decoding Site in Complex with Synthetic Aminoglycoside with F-Haba Group also contains other interesting chemical elements:
| Potassium | (K) | 1 atom |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of the Bacterial Ribosomal Decoding Site in Complex with Synthetic Aminoglycoside with F-Haba Group
(pdb code 3wru). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Crystal Structure of the Bacterial Ribosomal Decoding Site in Complex with Synthetic Aminoglycoside with F-Haba Group, PDB code: 3wru:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Crystal Structure of the Bacterial Ribosomal Decoding Site in Complex with Synthetic Aminoglycoside with F-Haba Group, PDB code: 3wru:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 3wru
Go back to
Fluorine binding site 1 out
of 2 in the Crystal Structure of the Bacterial Ribosomal Decoding Site in Complex with Synthetic Aminoglycoside with F-Haba Group
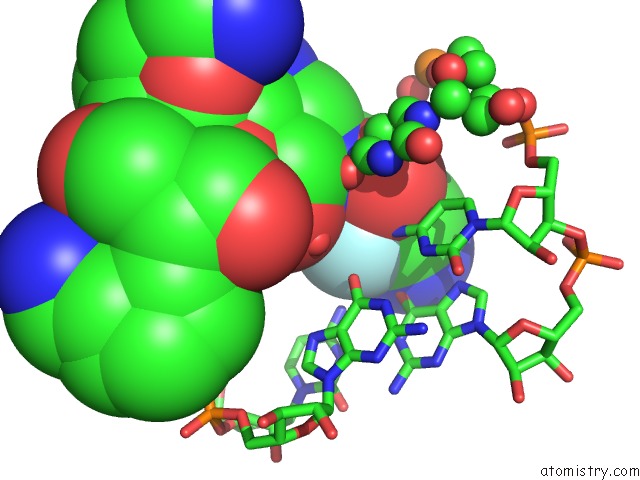
Mono view
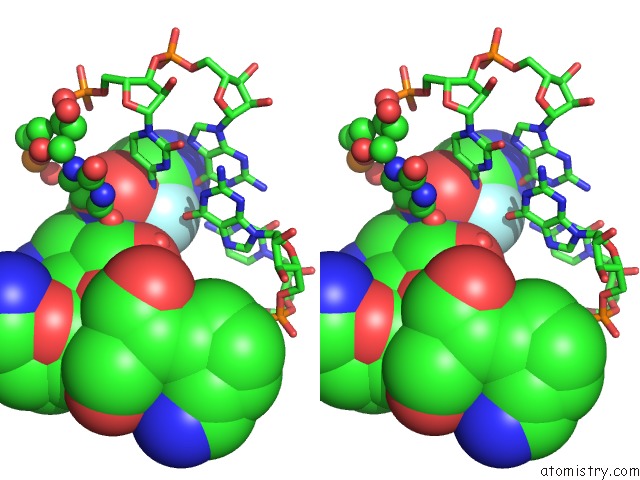
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of the Bacterial Ribosomal Decoding Site in Complex with Synthetic Aminoglycoside with F-Haba Group within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 3wru
Go back to
Fluorine binding site 2 out
of 2 in the Crystal Structure of the Bacterial Ribosomal Decoding Site in Complex with Synthetic Aminoglycoside with F-Haba Group
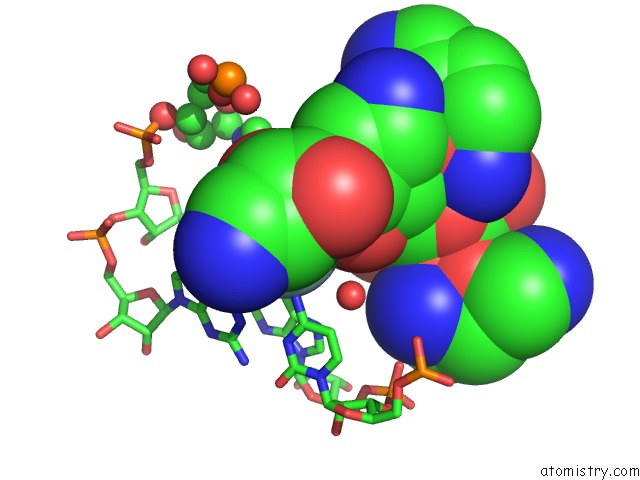
Mono view
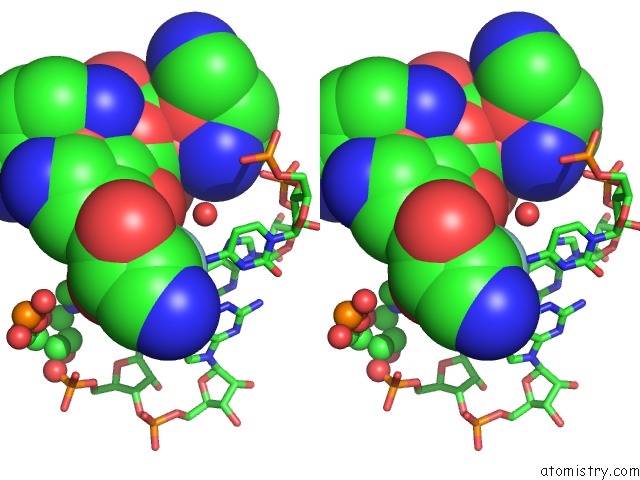
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of the Bacterial Ribosomal Decoding Site in Complex with Synthetic Aminoglycoside with F-Haba Group within 5.0Å range:
|
Reference:
J.P.Maianti,
H.Kanazawa,
P.Dozzo,
R.D.Matias,
L.A.Feeney,
E.S.Armstrong,
D.J.Hildebrandt,
T.R.Kane,
M.J.Gliedt,
A.A.Goldblum,
M.S.Linsell,
J.B.Aggen,
J.Kondo,
S.Hanessian.
Toxicity Modulation, Resistance Enzyme Evasion, and A-Site X-Ray Structure of Broad-Spectrum Antibacterial Neomycin Analogs Acs Chem.Biol. V. 9 2067 2014.
ISSN: ISSN 1554-8929
PubMed: 25019242
DOI: 10.1021/CB5003416
Page generated: Wed Jul 31 23:34:49 2024
ISSN: ISSN 1554-8929
PubMed: 25019242
DOI: 10.1021/CB5003416
Last articles
Ca in 5W0UCa in 5W78
Ca in 5W1F
Ca in 5W4S
Ca in 5W4C
Ca in 5W3Q
Ca in 5W2Y
Ca in 5W2W
Ca in 5W2U
Ca in 5W26