Fluorine »
PDB 3wyl-460d »
3znr »
Fluorine in PDB 3znr: HDAC7 Bound with Inhibitor TMP269
Enzymatic activity of HDAC7 Bound with Inhibitor TMP269
All present enzymatic activity of HDAC7 Bound with Inhibitor TMP269:
3.5.1.98;
3.5.1.98;
Protein crystallography data
The structure of HDAC7 Bound with Inhibitor TMP269, PDB code: 3znr
was solved by
M.Lobera,
K.Madauss,
D.Pohlhaus,
R.Trump,
M.Nolan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 70.49 / 2.40 |
| Space group | P 32 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.393, 81.393, 149.265, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 21.235 / 24.102 |
Other elements in 3znr:
The structure of HDAC7 Bound with Inhibitor TMP269 also contains other interesting chemical elements:
| Potassium | (K) | 6 atoms |
| Zinc | (Zn) | 6 atoms |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the HDAC7 Bound with Inhibitor TMP269
(pdb code 3znr). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the HDAC7 Bound with Inhibitor TMP269, PDB code: 3znr:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the HDAC7 Bound with Inhibitor TMP269, PDB code: 3znr:
Jump to Fluorine binding site number: 1; 2; 3;
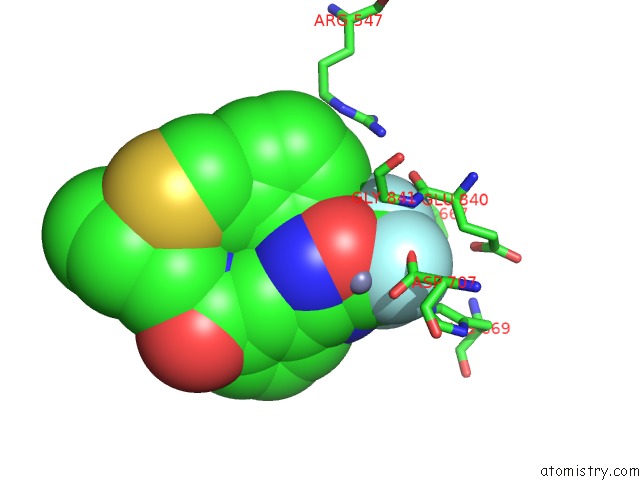
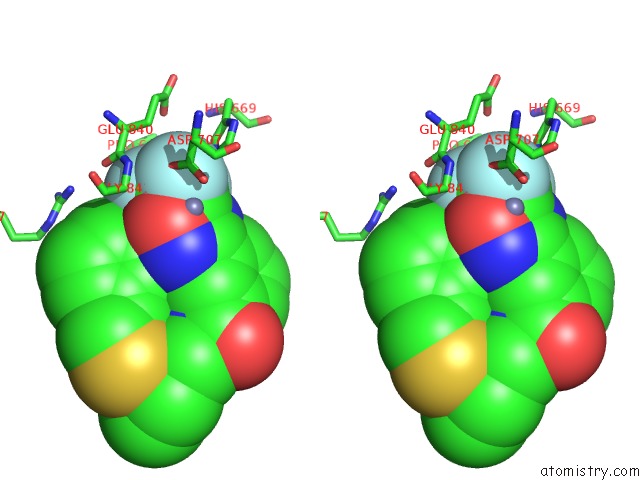
Fluorine binding site 1 out of 3 in 3znr
Go back to
Fluorine binding site 1 out
of 3 in the HDAC7 Bound with Inhibitor TMP269
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of HDAC7 Bound with Inhibitor TMP269 within 5.0Å range:
|
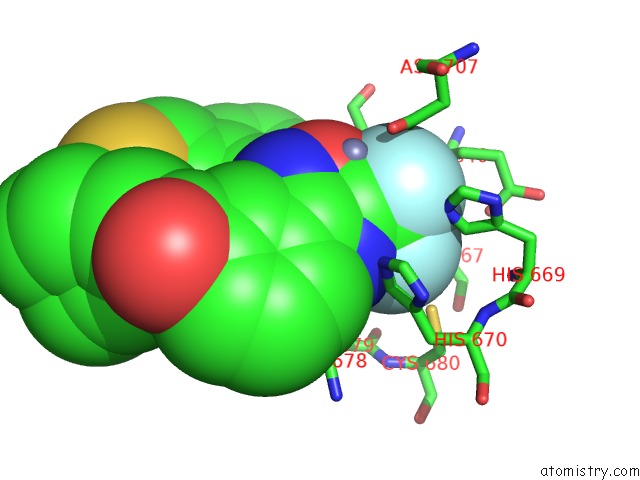
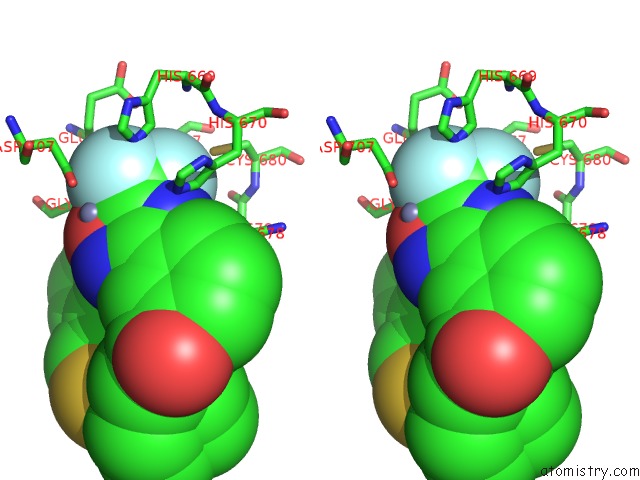
Fluorine binding site 2 out of 3 in 3znr
Go back to
Fluorine binding site 2 out
of 3 in the HDAC7 Bound with Inhibitor TMP269
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of HDAC7 Bound with Inhibitor TMP269 within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 3znr
Go back to
Fluorine binding site 3 out
of 3 in the HDAC7 Bound with Inhibitor TMP269
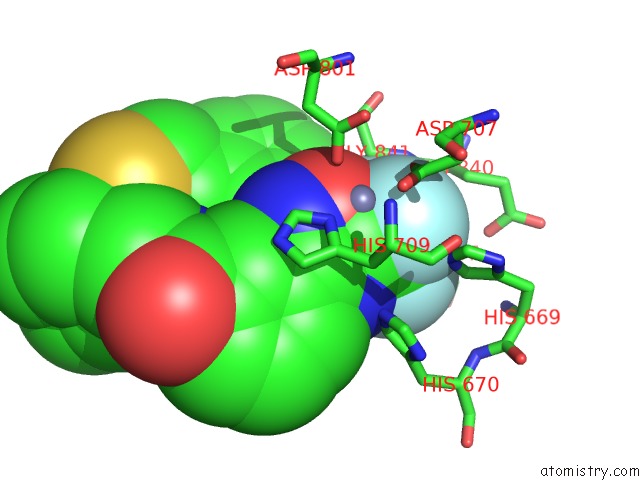
Mono view
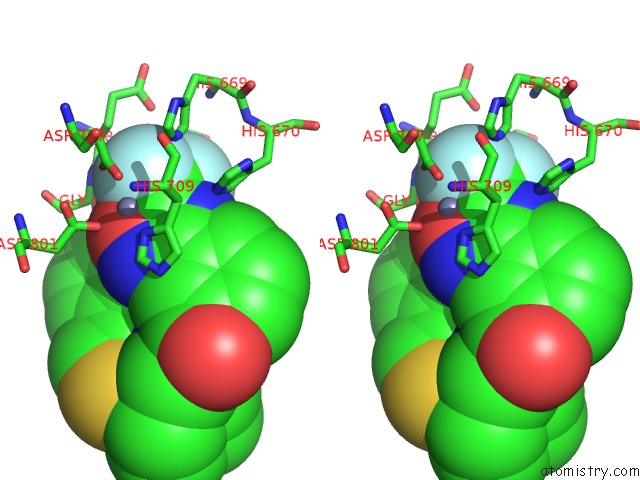
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of HDAC7 Bound with Inhibitor TMP269 within 5.0Å range:
|
Reference:
M.Lobera,
K.P.Madauss,
D.T.Pohlhaus,
Q.G.Wright,
M.Trocha,
D.R.Schmidt,
E.Baloglu,
R.P.Trump,
M.S.Head,
G.A.Hofmann,
M.Murray-Thompson,
B.Schwartz,
S.Chakravorty,
Z.Wu,
P.K.Mander,
L.Kruidenier,
R.A.Reid,
W.Burkhart,
B.J.Turunen,
J.X.Rong,
C.Wagner,
M.B.Moyer,
C.Wells,
X.Hong,
J.T.Moore,
J.D.Williams,
D.Soler,
S.Ghosh,
M.A.Nolan.
Selective Class Iia Histone Deacetylase Inhibition Via A Non-Chelating Zinc Binding Group Nat.Chem.Biol. V. 9 319 2013.
ISSN: ISSN 1552-4450
PubMed: 23524983
DOI: 10.1038/NCHEMBIO.1223
Page generated: Mon Jul 14 20:15:24 2025
ISSN: ISSN 1552-4450
PubMed: 23524983
DOI: 10.1038/NCHEMBIO.1223
Last articles
F in 4BS5F in 4BQR
F in 4BNT
F in 4BO3
F in 4BPM
F in 4BMM
F in 4BO8
F in 4BNK
F in 4BM8
F in 4BKZ