Fluorine »
PDB 3wyl-460d »
3zos »
Fluorine in PDB 3zos: Structure of the DDR1 Kinase Domain in Complex with Ponatinib
Enzymatic activity of Structure of the DDR1 Kinase Domain in Complex with Ponatinib
All present enzymatic activity of Structure of the DDR1 Kinase Domain in Complex with Ponatinib:
2.7.10.1;
2.7.10.1;
Protein crystallography data
The structure of Structure of the DDR1 Kinase Domain in Complex with Ponatinib, PDB code: 3zos
was solved by
P.Canning,
J.M.Elkins,
S.Goubin,
P.Mahajan,
A.Bradley,
D.Coutandin,
F.Vondelft,
C.H.Arrowsmith,
A.M.Edwards,
C.Bountra,
A.Bullock,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.41 / 1.92 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.951, 61.733, 80.193, 90.00, 104.40, 90.00 |
| R / Rfree (%) | 20.6 / 23.4 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib
(pdb code 3zos). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 9 binding sites of Fluorine where determined in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib, PDB code: 3zos:
Jump to Fluorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
In total 9 binding sites of Fluorine where determined in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib, PDB code: 3zos:
Jump to Fluorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
Fluorine binding site 1 out of 9 in 3zos
Go back to
Fluorine binding site 1 out
of 9 in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib
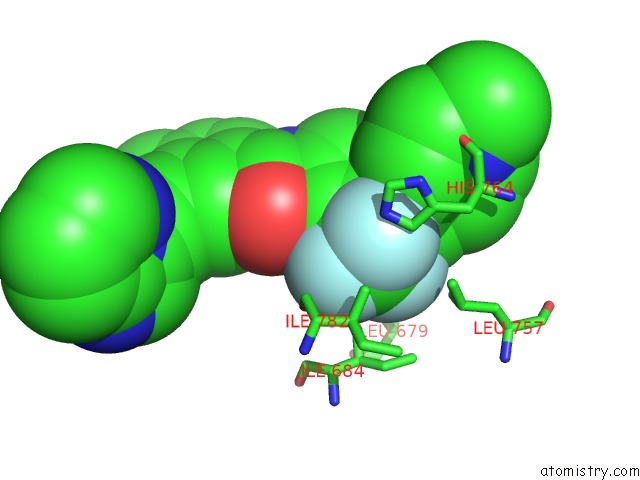
Mono view
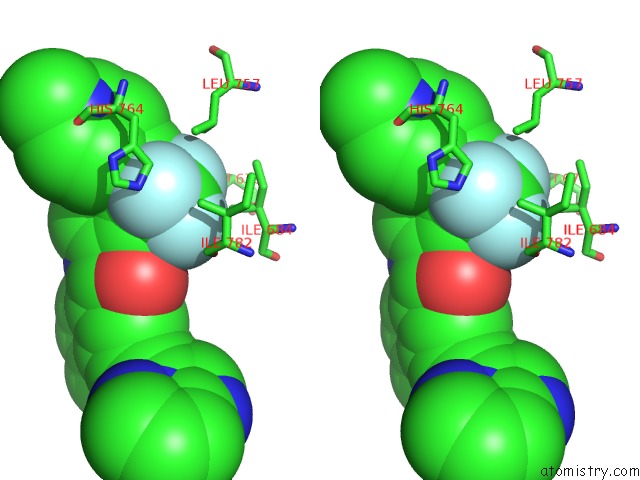
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Structure of the DDR1 Kinase Domain in Complex with Ponatinib within 5.0Å range:
|
Fluorine binding site 2 out of 9 in 3zos
Go back to
Fluorine binding site 2 out
of 9 in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib
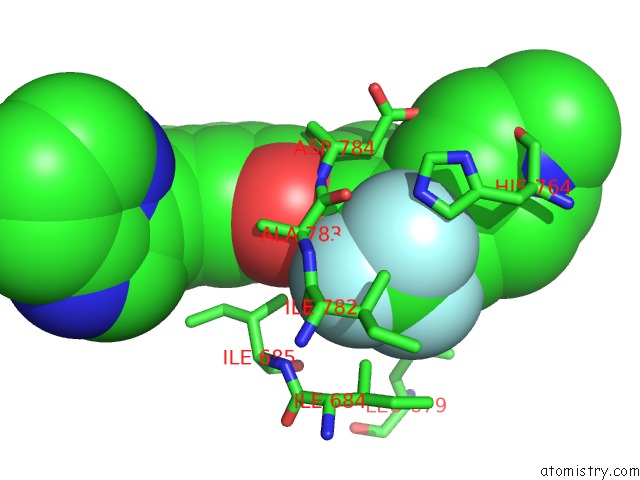
Mono view
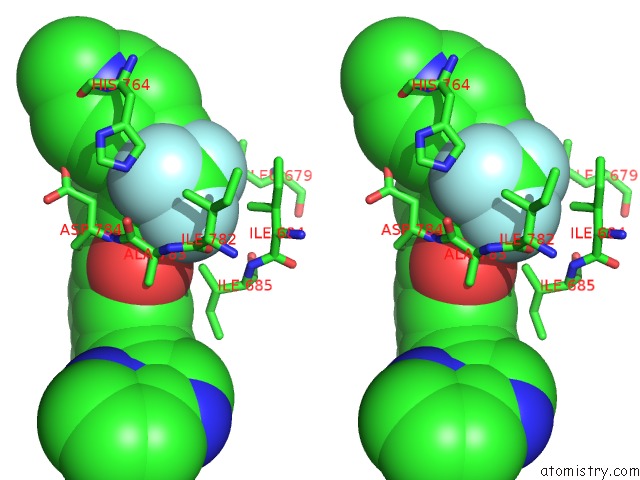
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Structure of the DDR1 Kinase Domain in Complex with Ponatinib within 5.0Å range:
|
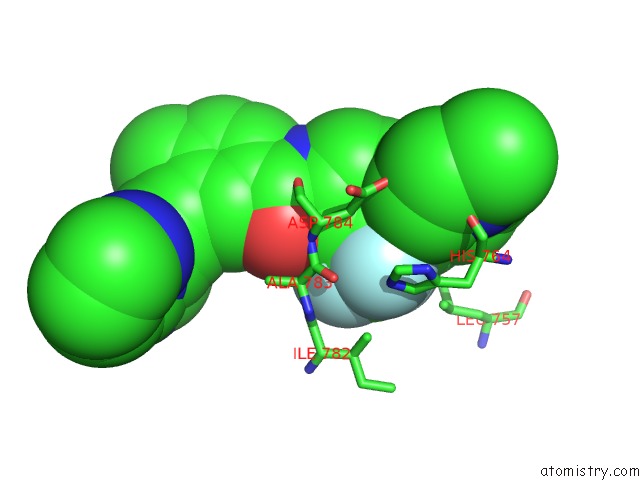
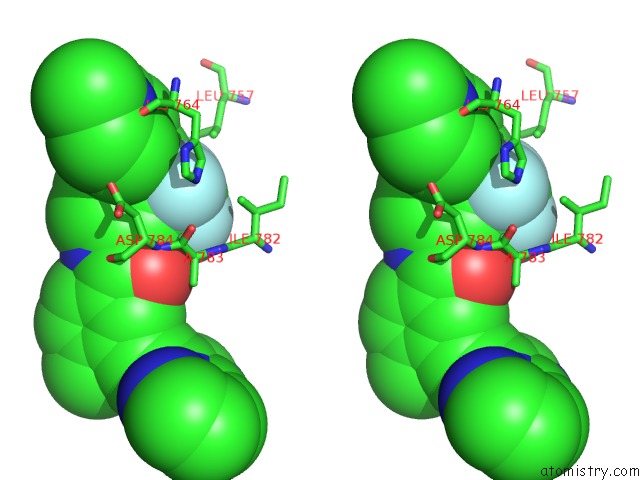
Fluorine binding site 3 out of 9 in 3zos
Go back to
Fluorine binding site 3 out
of 9 in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Structure of the DDR1 Kinase Domain in Complex with Ponatinib within 5.0Å range:
|
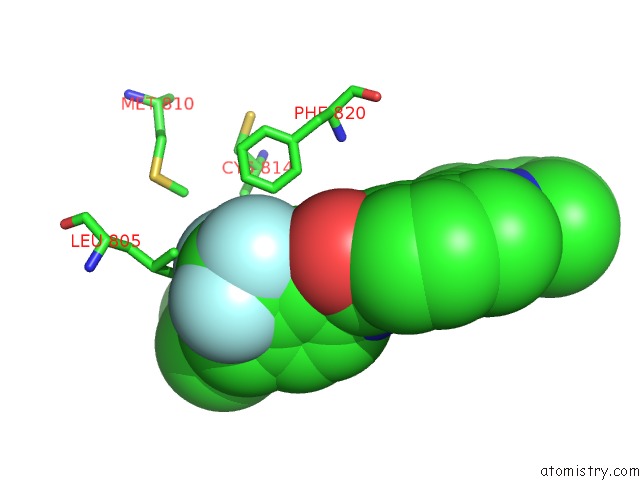
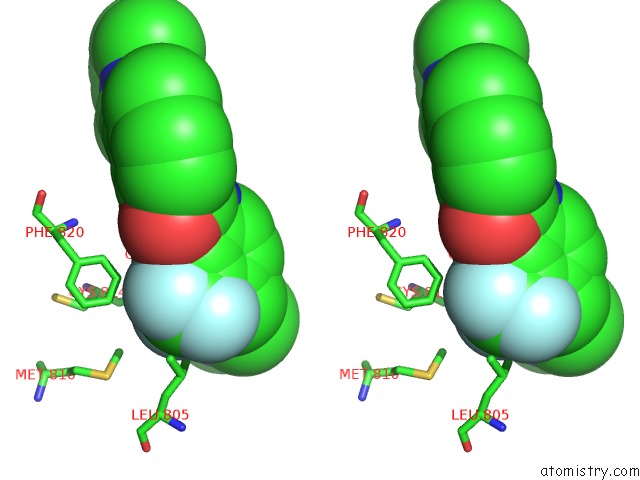
Fluorine binding site 4 out of 9 in 3zos
Go back to
Fluorine binding site 4 out
of 9 in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 4 of Structure of the DDR1 Kinase Domain in Complex with Ponatinib within 5.0Å range:
|
Fluorine binding site 5 out of 9 in 3zos
Go back to
Fluorine binding site 5 out
of 9 in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib
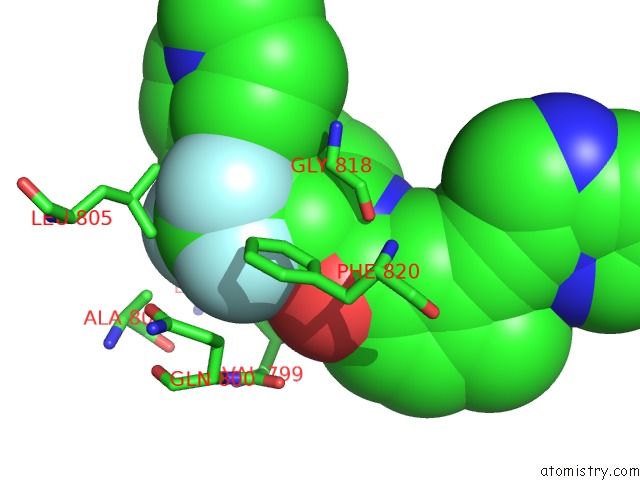
Mono view
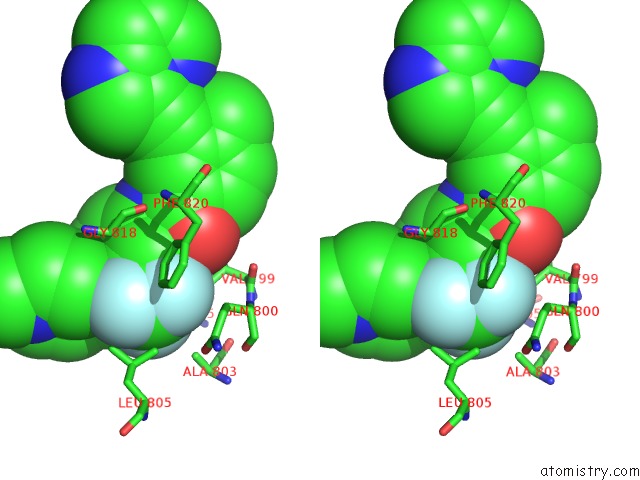
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 5 of Structure of the DDR1 Kinase Domain in Complex with Ponatinib within 5.0Å range:
|
Fluorine binding site 6 out of 9 in 3zos
Go back to
Fluorine binding site 6 out
of 9 in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib
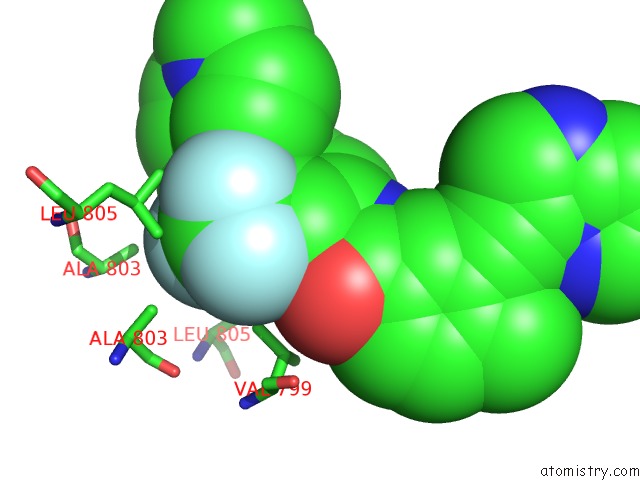
Mono view
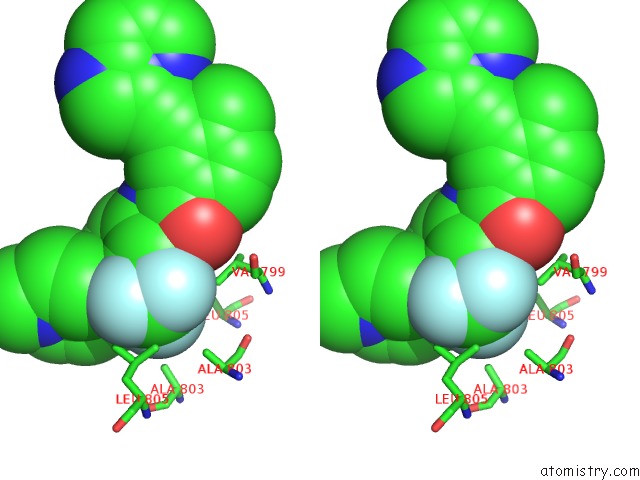
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 6 of Structure of the DDR1 Kinase Domain in Complex with Ponatinib within 5.0Å range:
|
Fluorine binding site 7 out of 9 in 3zos
Go back to
Fluorine binding site 7 out
of 9 in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib
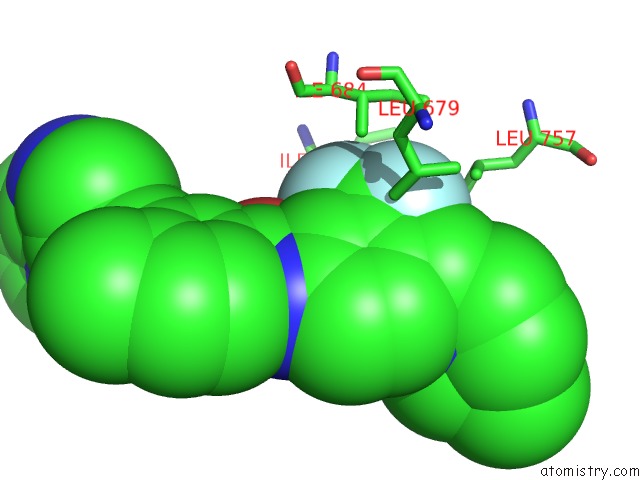
Mono view
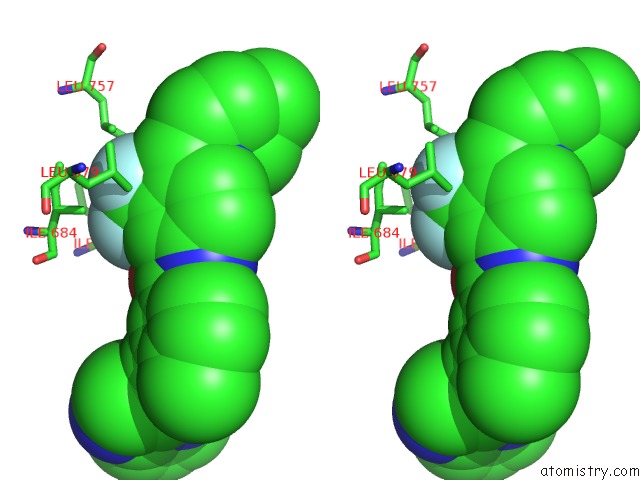
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 7 of Structure of the DDR1 Kinase Domain in Complex with Ponatinib within 5.0Å range:
|
Fluorine binding site 8 out of 9 in 3zos
Go back to
Fluorine binding site 8 out
of 9 in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib
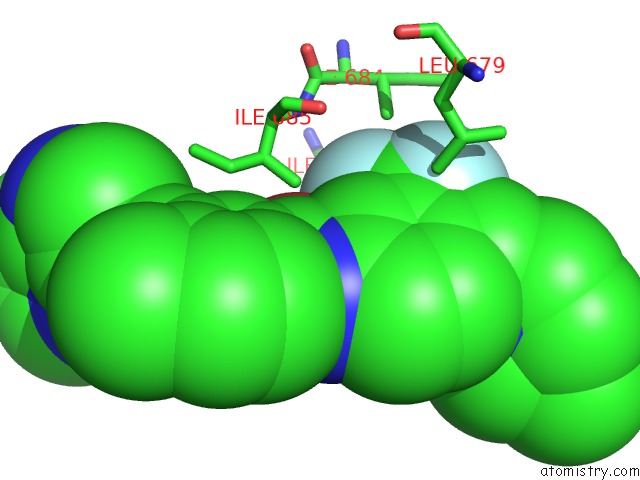
Mono view
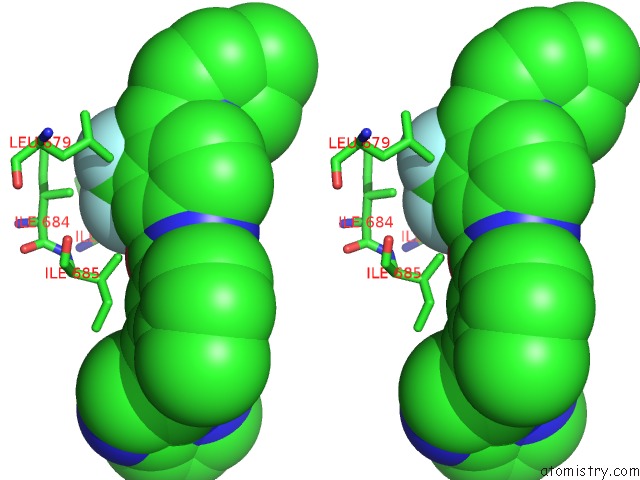
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 8 of Structure of the DDR1 Kinase Domain in Complex with Ponatinib within 5.0Å range:
|
Fluorine binding site 9 out of 9 in 3zos
Go back to
Fluorine binding site 9 out
of 9 in the Structure of the DDR1 Kinase Domain in Complex with Ponatinib
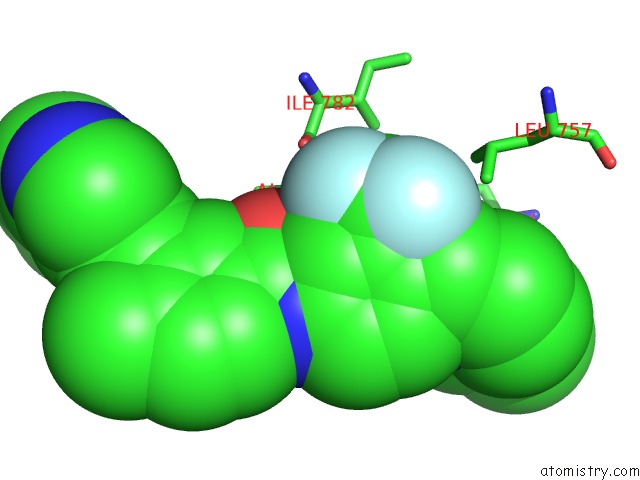
Mono view
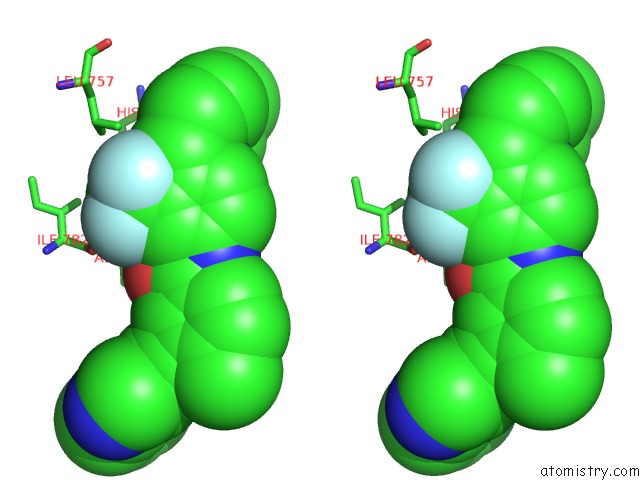
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 9 of Structure of the DDR1 Kinase Domain in Complex with Ponatinib within 5.0Å range:
|
Reference:
P.Canning,
J.M.Elkins,
S.Goubin,
P.Mahajan,
A.Bradley,
D.Coutandin,
F.Von Delft,
C.H.Arrowsmith,
A.M.Edwards,
C.Bountra,
A.Bullock.
Structural Mechanisms Determining Inhibition of the Collagen Receptor DDR1 By Selective and Multi-Targeted Type II Kinase Inhibitors J.Mol.Biol. V. 426 2457 2014.
ISSN: ISSN 0022-2836
PubMed: 24768818
DOI: 10.1016/J.JMB.2014.04.014
Page generated: Wed Jul 31 23:47:42 2024
ISSN: ISSN 0022-2836
PubMed: 24768818
DOI: 10.1016/J.JMB.2014.04.014
Last articles
Cl in 7TNMCl in 7TNN
Cl in 7TNL
Cl in 7TNK
Cl in 7TNJ
Cl in 7TNH
Cl in 7TNF
Cl in 7TND
Cl in 7TN8
Cl in 7TNC