Fluorine »
PDB 4gpb-4hw7 »
4gs0 »
Fluorine in PDB 4gs0: Crystal Structure of SHP1 Catalytic Domain with JAK1 Activation Loop Peptide
Enzymatic activity of Crystal Structure of SHP1 Catalytic Domain with JAK1 Activation Loop Peptide
All present enzymatic activity of Crystal Structure of SHP1 Catalytic Domain with JAK1 Activation Loop Peptide:
2.7.10.2; 3.1.3.48;
2.7.10.2; 3.1.3.48;
Protein crystallography data
The structure of Crystal Structure of SHP1 Catalytic Domain with JAK1 Activation Loop Peptide, PDB code: 4gs0
was solved by
N.L.Alicea-Velazquez,
J.Jakoncic,
T.J.Boggon,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.02 / 1.80 |
| Space group | P 31 |
| Cell size a, b, c (Å), α, β, γ (°) | 43.935, 43.935, 258.112, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 16.2 / 19.5 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of SHP1 Catalytic Domain with JAK1 Activation Loop Peptide
(pdb code 4gs0). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Crystal Structure of SHP1 Catalytic Domain with JAK1 Activation Loop Peptide, PDB code: 4gs0:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Crystal Structure of SHP1 Catalytic Domain with JAK1 Activation Loop Peptide, PDB code: 4gs0:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 4gs0
Go back to
Fluorine binding site 1 out
of 2 in the Crystal Structure of SHP1 Catalytic Domain with JAK1 Activation Loop Peptide
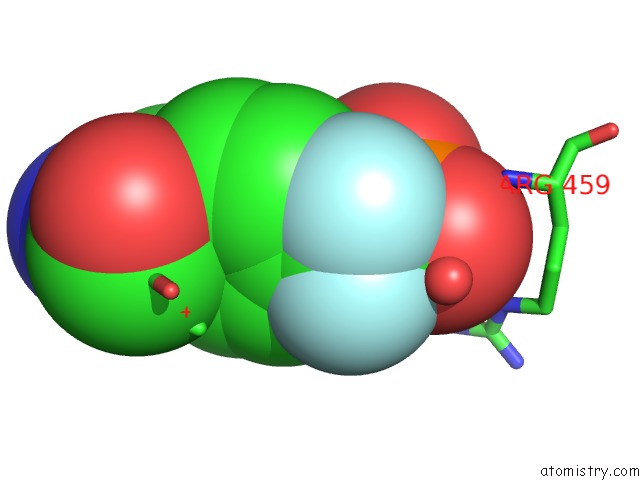
Mono view
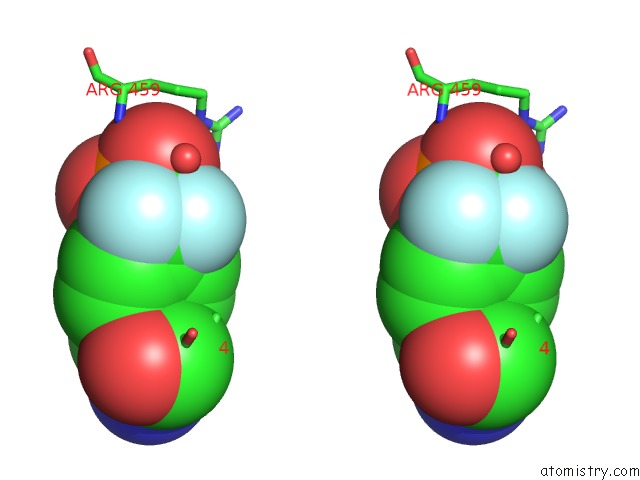
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of SHP1 Catalytic Domain with JAK1 Activation Loop Peptide within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 4gs0
Go back to
Fluorine binding site 2 out
of 2 in the Crystal Structure of SHP1 Catalytic Domain with JAK1 Activation Loop Peptide
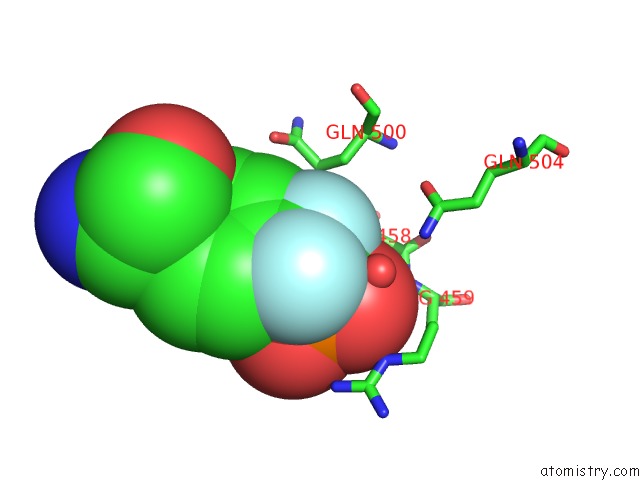
Mono view
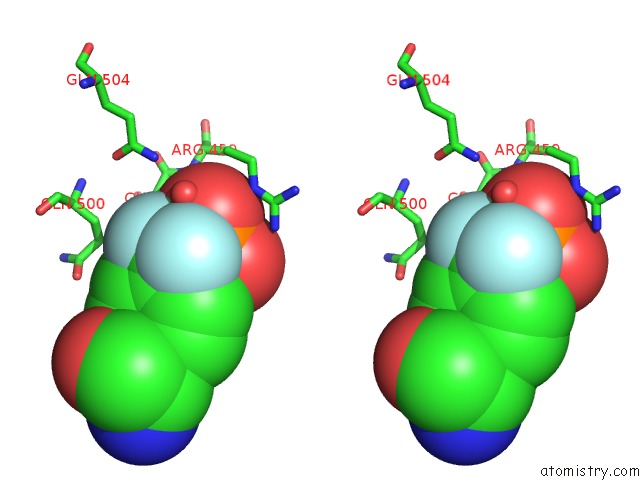
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of SHP1 Catalytic Domain with JAK1 Activation Loop Peptide within 5.0Å range:
|
Reference:
N.L.Alicea-Velazquez,
J.Jakoncic,
T.J.Boggon.
Structure-Guided Studies of the Shp-1/JAK1 Interaction Provide New Insights Into Phosphatase Catalytic Domain Substrate Recognition. J.Struct.Biol. V. 181 243 2013.
ISSN: ISSN 1047-8477
PubMed: 23296072
DOI: 10.1016/J.JSB.2012.12.009
Page generated: Thu Aug 1 02:00:54 2024
ISSN: ISSN 1047-8477
PubMed: 23296072
DOI: 10.1016/J.JSB.2012.12.009
Last articles
Cl in 7T8KCl in 7T8L
Cl in 7T80
Cl in 7T8J
Cl in 7T88
Cl in 7T4W
Cl in 7T4V
Cl in 7T7K
Cl in 7T6S
Cl in 7T6C