Fluorine »
PDB 4gpb-4hw7 »
4gvh »
Fluorine in PDB 4gvh: Crystal Structure of Salmonella Typhimurium Family 3 Glycoside Hydrolase (Nagz) Covalently Bound to 5-Fluoro-Glcnac.
Enzymatic activity of Crystal Structure of Salmonella Typhimurium Family 3 Glycoside Hydrolase (Nagz) Covalently Bound to 5-Fluoro-Glcnac.
All present enzymatic activity of Crystal Structure of Salmonella Typhimurium Family 3 Glycoside Hydrolase (Nagz) Covalently Bound to 5-Fluoro-Glcnac.:
3.2.1.52;
3.2.1.52;
Protein crystallography data
The structure of Crystal Structure of Salmonella Typhimurium Family 3 Glycoside Hydrolase (Nagz) Covalently Bound to 5-Fluoro-Glcnac., PDB code: 4gvh
was solved by
J.P.Bacik,
B.L.Mark,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.62 / 1.45 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.260, 65.923, 94.724, 90.00, 99.28, 90.00 |
| R / Rfree (%) | 16.5 / 19.5 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of Salmonella Typhimurium Family 3 Glycoside Hydrolase (Nagz) Covalently Bound to 5-Fluoro-Glcnac.
(pdb code 4gvh). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Crystal Structure of Salmonella Typhimurium Family 3 Glycoside Hydrolase (Nagz) Covalently Bound to 5-Fluoro-Glcnac., PDB code: 4gvh:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Crystal Structure of Salmonella Typhimurium Family 3 Glycoside Hydrolase (Nagz) Covalently Bound to 5-Fluoro-Glcnac., PDB code: 4gvh:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 4gvh
Go back to
Fluorine binding site 1 out
of 2 in the Crystal Structure of Salmonella Typhimurium Family 3 Glycoside Hydrolase (Nagz) Covalently Bound to 5-Fluoro-Glcnac.
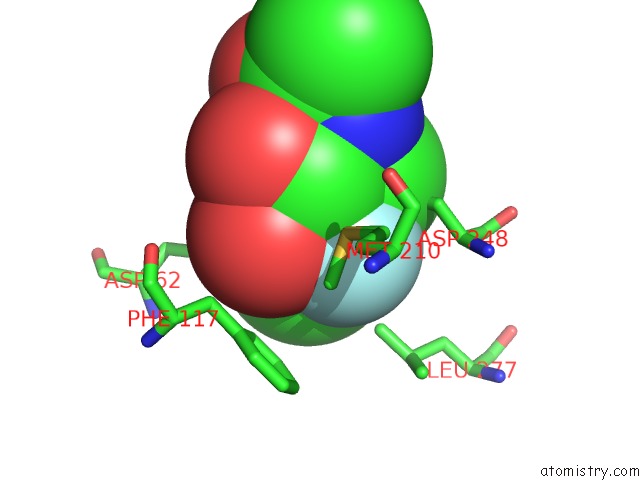
Mono view
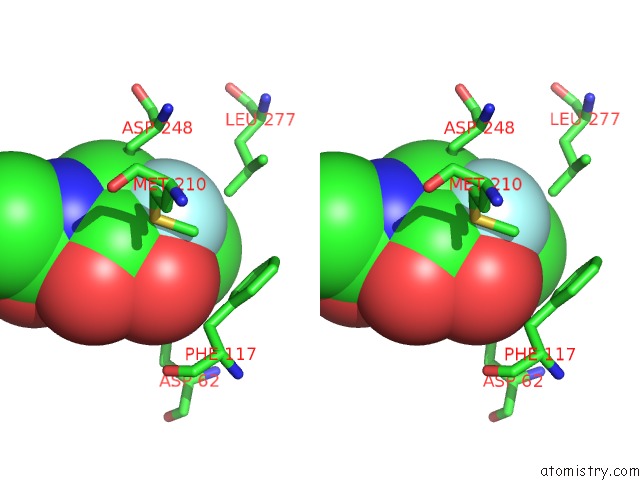
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of Salmonella Typhimurium Family 3 Glycoside Hydrolase (Nagz) Covalently Bound to 5-Fluoro-Glcnac. within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 4gvh
Go back to
Fluorine binding site 2 out
of 2 in the Crystal Structure of Salmonella Typhimurium Family 3 Glycoside Hydrolase (Nagz) Covalently Bound to 5-Fluoro-Glcnac.
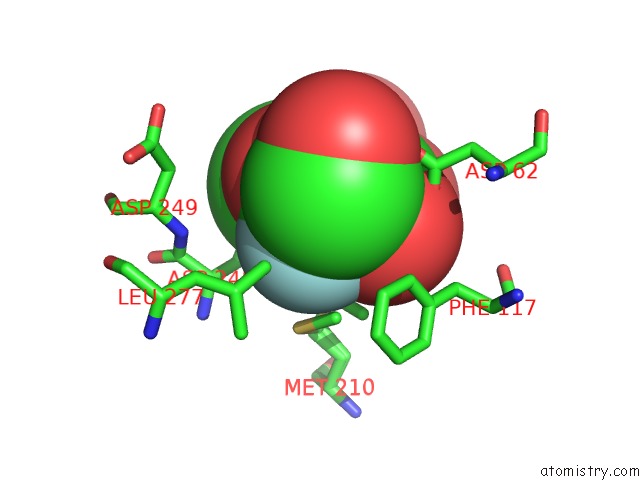
Mono view
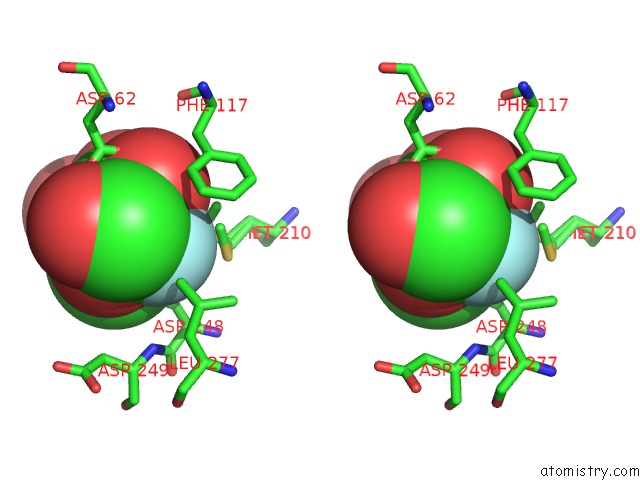
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of Salmonella Typhimurium Family 3 Glycoside Hydrolase (Nagz) Covalently Bound to 5-Fluoro-Glcnac. within 5.0Å range:
|
Reference:
J.P.Bacik,
G.E.Whitworth,
K.A.Stubbs,
D.J.Vocadlo,
B.L.Mark.
Active Site Plasticity Within the Glycoside Hydrolase Nagz Underlies A Dynamic Mechanism of Substrate Distortion. Chem.Biol. V. 19 1471 2012.
ISSN: ISSN 1074-5521
PubMed: 23177201
DOI: 10.1016/J.CHEMBIOL.2012.09.016
Page generated: Thu Aug 1 02:00:54 2024
ISSN: ISSN 1074-5521
PubMed: 23177201
DOI: 10.1016/J.CHEMBIOL.2012.09.016
Last articles
Ca in 5Y7GCa in 5Y8A
Ca in 5Y31
Ca in 5Y7F
Ca in 5Y5E
Ca in 5Y4F
Ca in 5Y17
Ca in 5XZN
Ca in 5XZM
Ca in 5XY4