Fluorine »
PDB 4iju-4j0t »
4ivo »
Fluorine in PDB 4ivo: Structure of Human Protoporphyrinogen IX Oxidase(R59Q)
Enzymatic activity of Structure of Human Protoporphyrinogen IX Oxidase(R59Q)
All present enzymatic activity of Structure of Human Protoporphyrinogen IX Oxidase(R59Q):
1.3.3.4;
1.3.3.4;
Protein crystallography data
The structure of Structure of Human Protoporphyrinogen IX Oxidase(R59Q), PDB code: 4ivo
was solved by
Q.Xiaohong,
W.Baifan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.58 / 2.60 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 135.809, 135.809, 158.338, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 17.7 / 23.8 |
Other elements in 4ivo:
The structure of Structure of Human Protoporphyrinogen IX Oxidase(R59Q) also contains other interesting chemical elements:
| Chlorine | (Cl) | 1 atom |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Structure of Human Protoporphyrinogen IX Oxidase(R59Q)
(pdb code 4ivo). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Structure of Human Protoporphyrinogen IX Oxidase(R59Q), PDB code: 4ivo:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Structure of Human Protoporphyrinogen IX Oxidase(R59Q), PDB code: 4ivo:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 4ivo
Go back to
Fluorine binding site 1 out
of 3 in the Structure of Human Protoporphyrinogen IX Oxidase(R59Q)
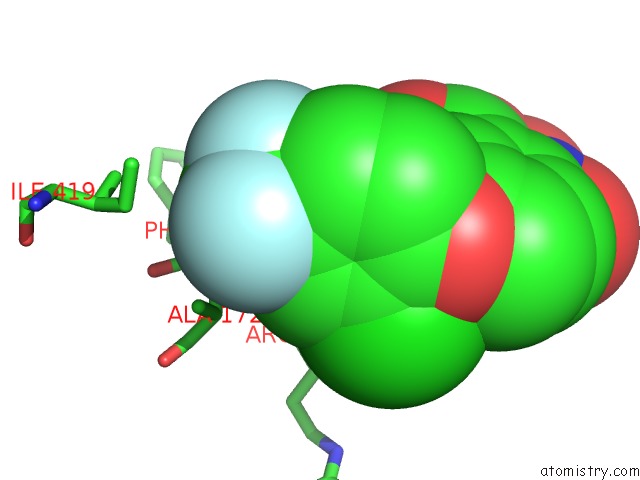
Mono view
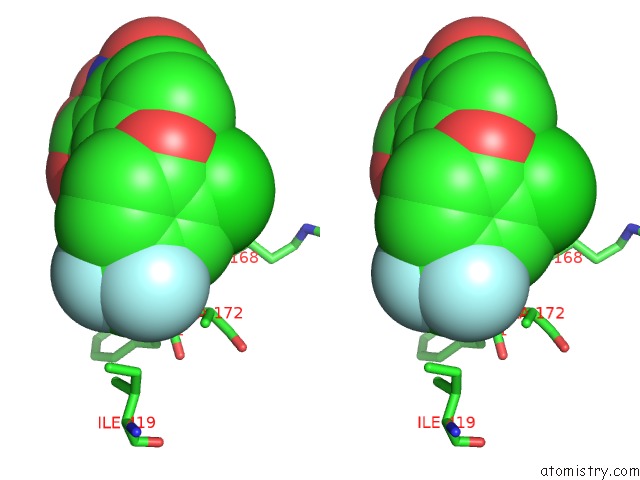
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Structure of Human Protoporphyrinogen IX Oxidase(R59Q) within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 4ivo
Go back to
Fluorine binding site 2 out
of 3 in the Structure of Human Protoporphyrinogen IX Oxidase(R59Q)
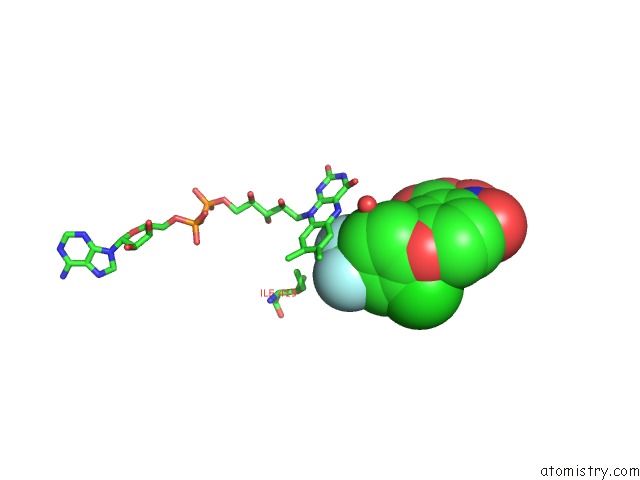
Mono view
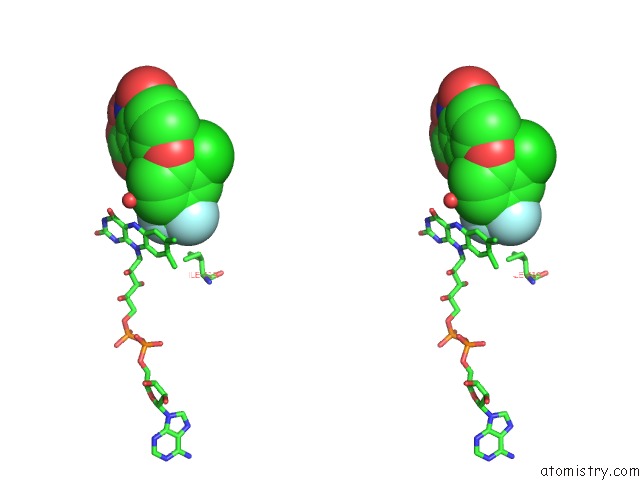
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Structure of Human Protoporphyrinogen IX Oxidase(R59Q) within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 4ivo
Go back to
Fluorine binding site 3 out
of 3 in the Structure of Human Protoporphyrinogen IX Oxidase(R59Q)
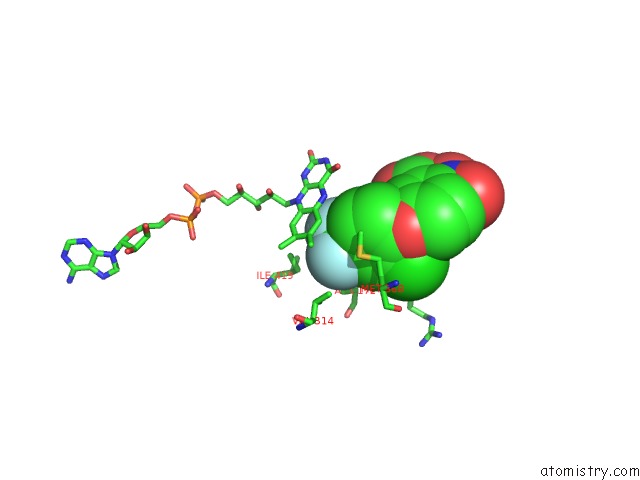
Mono view
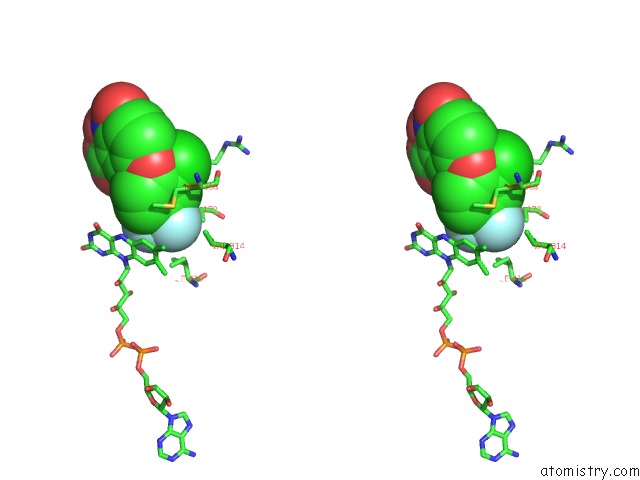
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Structure of Human Protoporphyrinogen IX Oxidase(R59Q) within 5.0Å range:
|
Reference:
B.Wang,
X.Wen,
X.Qin,
Z.Wang,
Y.Tan,
Y.Shen,
Z.Xi.
Quantitative Structural Insight Into Human Variegate Porphyria Disease. J.Biol.Chem. V. 288 11731 2013.
ISSN: ISSN 0021-9258
PubMed: 23467411
DOI: 10.1074/JBC.M113.459768
Page generated: Thu Aug 1 02:35:48 2024
ISSN: ISSN 0021-9258
PubMed: 23467411
DOI: 10.1074/JBC.M113.459768
Last articles
Cl in 5WKECl in 5WKG
Cl in 5WHV
Cl in 5WJL
Cl in 5WJK
Cl in 5WJI
Cl in 5WIV
Cl in 5WJ8
Cl in 5WII
Cl in 5WIU