Fluorine »
PDB 5hu9-5ifd »
5idp »
Fluorine in PDB 5idp: CDK8-Cycc in Complex with (3-Amino-1H-Indazol-5-Yl)-[(S)-2-(4-Fluoro- Phenyl)-Piperidin-1-Yl]-Methanone
Enzymatic activity of CDK8-Cycc in Complex with (3-Amino-1H-Indazol-5-Yl)-[(S)-2-(4-Fluoro- Phenyl)-Piperidin-1-Yl]-Methanone
All present enzymatic activity of CDK8-Cycc in Complex with (3-Amino-1H-Indazol-5-Yl)-[(S)-2-(4-Fluoro- Phenyl)-Piperidin-1-Yl]-Methanone:
2.7.11.22; 2.7.11.23;
2.7.11.22; 2.7.11.23;
Protein crystallography data
The structure of CDK8-Cycc in Complex with (3-Amino-1H-Indazol-5-Yl)-[(S)-2-(4-Fluoro- Phenyl)-Piperidin-1-Yl]-Methanone, PDB code: 5idp
was solved by
D.Musil,
J.Blagg,
A.Mallinger,
P.Czodrowski,
K.Schiemann,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 86.17 / 2.65 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 71.113, 71.093, 172.335, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.1 / 27.2 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the CDK8-Cycc in Complex with (3-Amino-1H-Indazol-5-Yl)-[(S)-2-(4-Fluoro- Phenyl)-Piperidin-1-Yl]-Methanone
(pdb code 5idp). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total only one binding site of Fluorine was determined in the CDK8-Cycc in Complex with (3-Amino-1H-Indazol-5-Yl)-[(S)-2-(4-Fluoro- Phenyl)-Piperidin-1-Yl]-Methanone, PDB code: 5idp:
In total only one binding site of Fluorine was determined in the CDK8-Cycc in Complex with (3-Amino-1H-Indazol-5-Yl)-[(S)-2-(4-Fluoro- Phenyl)-Piperidin-1-Yl]-Methanone, PDB code: 5idp:
Fluorine binding site 1 out of 1 in 5idp
Go back to
Fluorine binding site 1 out
of 1 in the CDK8-Cycc in Complex with (3-Amino-1H-Indazol-5-Yl)-[(S)-2-(4-Fluoro- Phenyl)-Piperidin-1-Yl]-Methanone
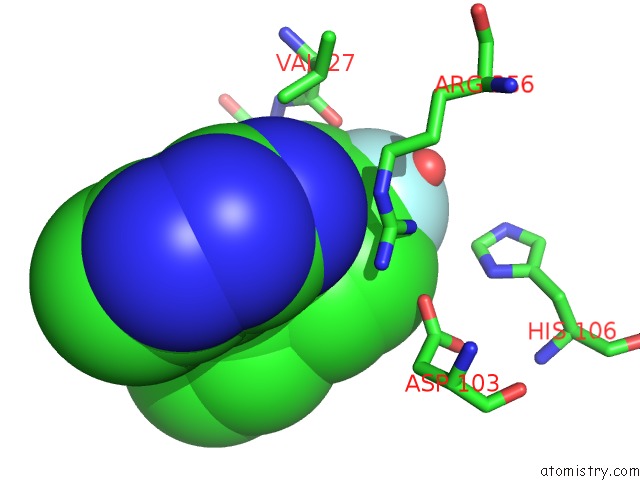
Mono view
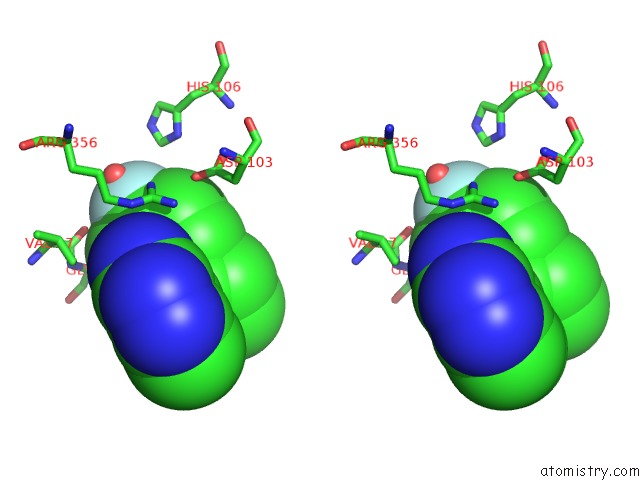
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of CDK8-Cycc in Complex with (3-Amino-1H-Indazol-5-Yl)-[(S)-2-(4-Fluoro- Phenyl)-Piperidin-1-Yl]-Methanone within 5.0Å range:
|
Reference:
P.Czodrowski,
A.Mallinger,
D.Wienke,
C.Esdar,
O.Poschke,
M.Busch,
F.Rohdich,
S.A.Eccles,
M.J.Ortiz-Ruiz,
R.Schneider,
F.I.Raynaud,
P.A.Clarke,
D.Musil,
D.Schwarz,
T.Dale,
K.Urbahns,
J.Blagg,
K.Schiemann.
Structure-Based Optimization of Potent, Selective, and Orally Bioavailable CDK8 Inhibitors Discovered By High-Throughput Screening. J. Med. Chem. V. 59 9337 2016.
ISSN: ISSN 1520-4804
PubMed: 27490956
DOI: 10.1021/ACS.JMEDCHEM.6B00597
Page generated: Tue Jul 15 04:08:31 2025
ISSN: ISSN 1520-4804
PubMed: 27490956
DOI: 10.1021/ACS.JMEDCHEM.6B00597
Last articles
F in 5Z3UF in 5Z5S
F in 5Z3V
F in 5Z12
F in 5YVO
F in 5YY1
F in 5Z1I
F in 5Z1H
F in 5YXZ
F in 5YWI