Fluorine »
PDB 5sjr-5sog »
5slq »
Fluorine in PDB 5slq: Pandda Analysis Group Deposition -- Crystal Structure of Sars-Cov-2 NSP14 in Complex with Z2856434829
Protein crystallography data
The structure of Pandda Analysis Group Deposition -- Crystal Structure of Sars-Cov-2 NSP14 in Complex with Z2856434829, PDB code: 5slq
was solved by
N.Imprachim,
Y.Yosaatmadja,
F.Von-Delft,
C.Bountra,
O.Gileadi,
J.A.Newman,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.90 / 2.11 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 67.795, 68.648, 138.407, 90, 90, 90 |
| R / Rfree (%) | 23.6 / 27.4 |
Other elements in 5slq:
The structure of Pandda Analysis Group Deposition -- Crystal Structure of Sars-Cov-2 NSP14 in Complex with Z2856434829 also contains other interesting chemical elements:
| Zinc | (Zn) | 3 atoms |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Pandda Analysis Group Deposition -- Crystal Structure of Sars-Cov-2 NSP14 in Complex with Z2856434829
(pdb code 5slq). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Pandda Analysis Group Deposition -- Crystal Structure of Sars-Cov-2 NSP14 in Complex with Z2856434829, PDB code: 5slq:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Pandda Analysis Group Deposition -- Crystal Structure of Sars-Cov-2 NSP14 in Complex with Z2856434829, PDB code: 5slq:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 5slq
Go back to
Fluorine binding site 1 out
of 2 in the Pandda Analysis Group Deposition -- Crystal Structure of Sars-Cov-2 NSP14 in Complex with Z2856434829
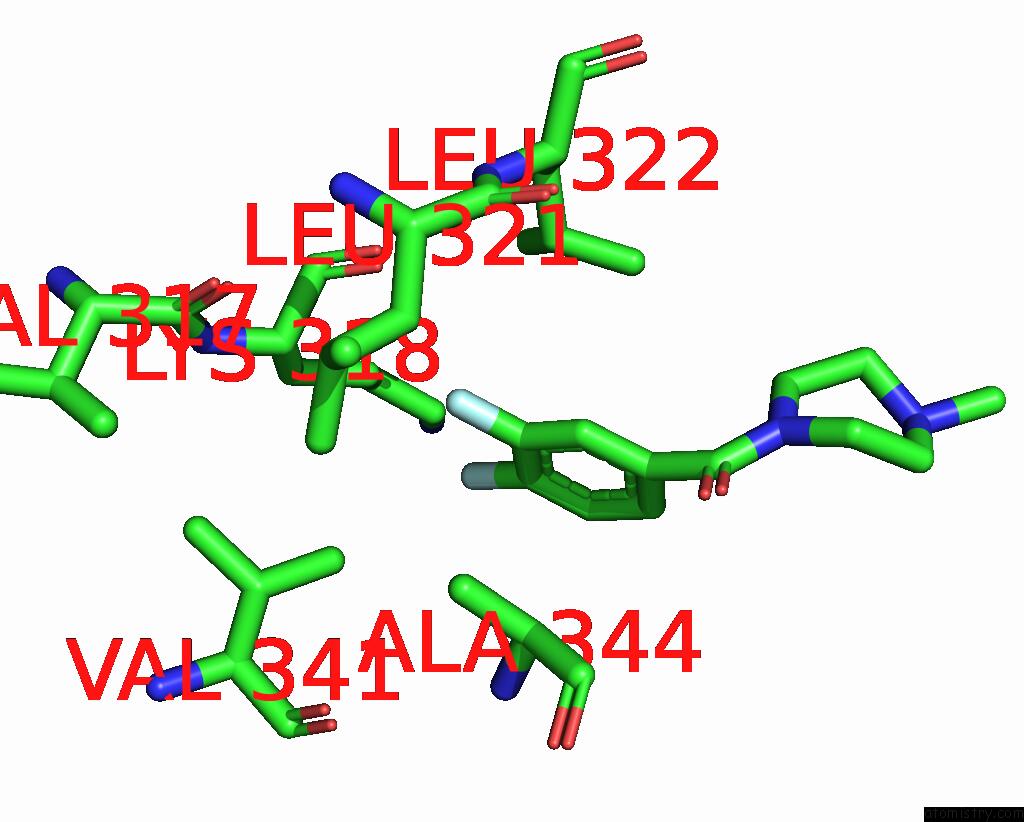
Mono view
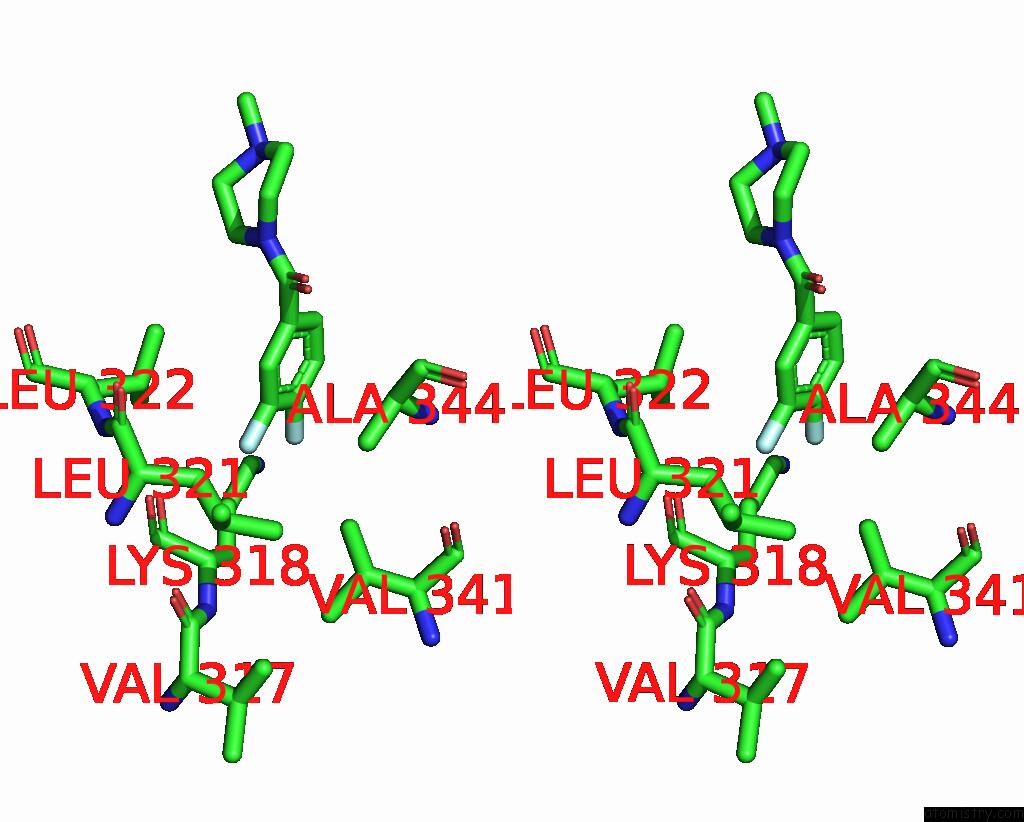
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Pandda Analysis Group Deposition -- Crystal Structure of Sars-Cov-2 NSP14 in Complex with Z2856434829 within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 5slq
Go back to
Fluorine binding site 2 out
of 2 in the Pandda Analysis Group Deposition -- Crystal Structure of Sars-Cov-2 NSP14 in Complex with Z2856434829
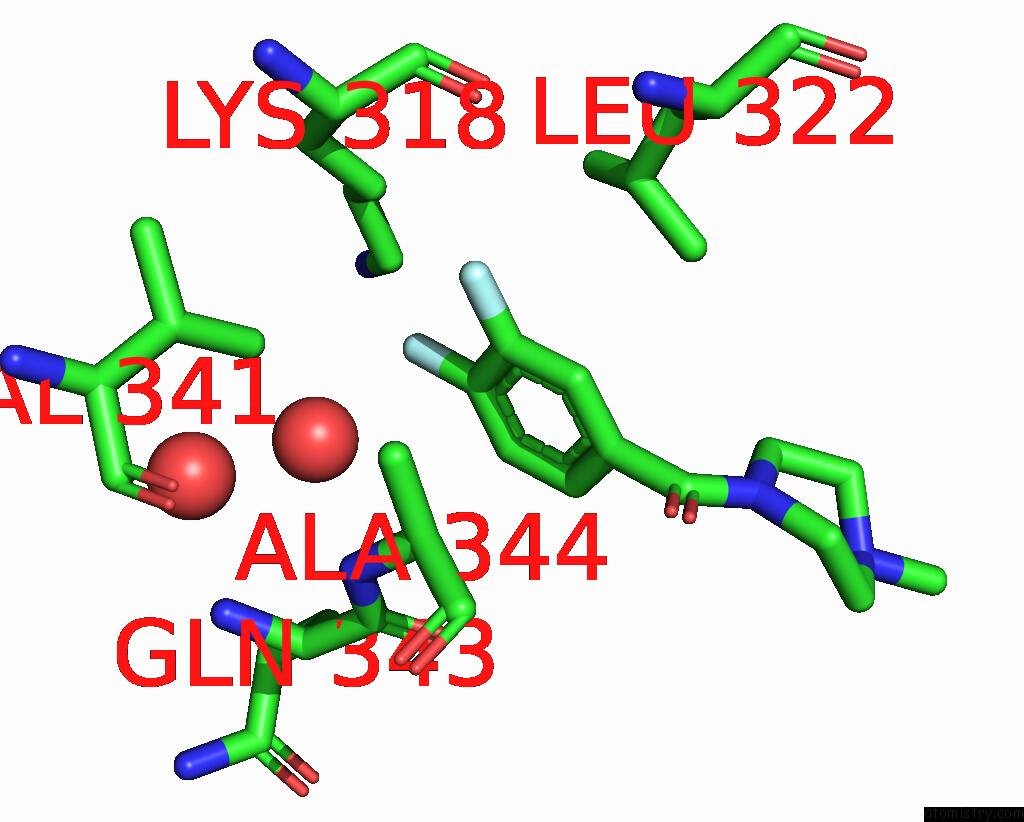
Mono view
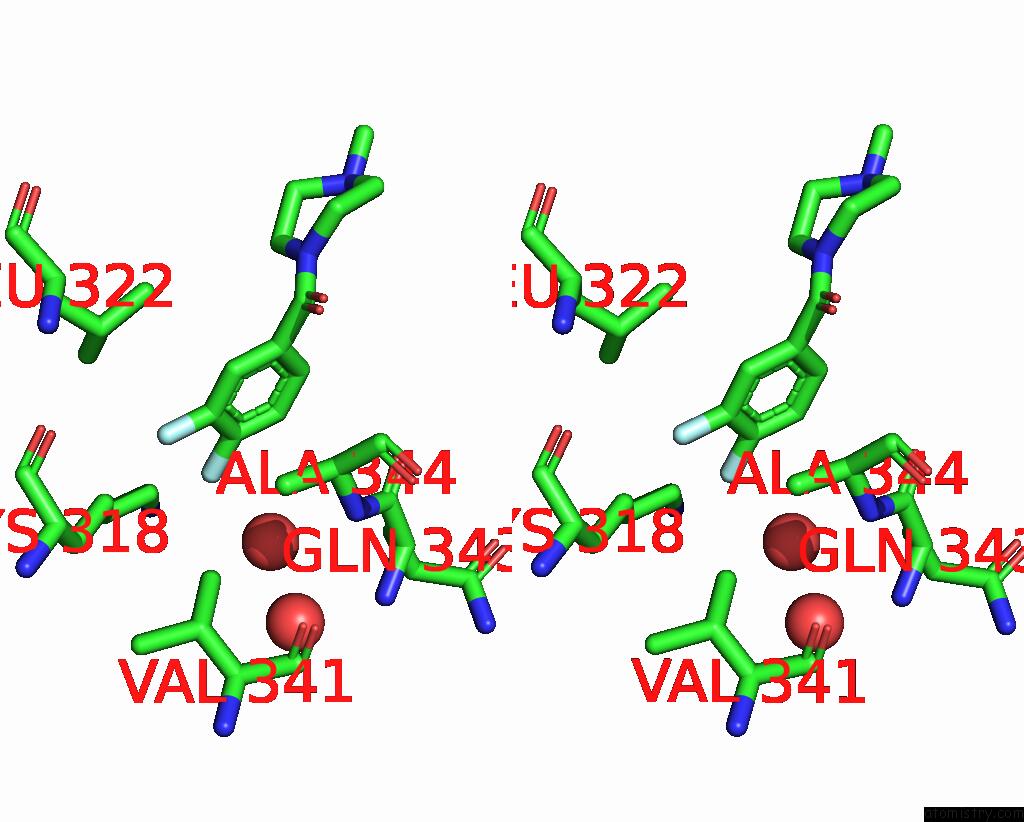
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Pandda Analysis Group Deposition -- Crystal Structure of Sars-Cov-2 NSP14 in Complex with Z2856434829 within 5.0Å range:
|
Reference:
N.Imprachim,
Y.Yosaatmadja,
F.Von-Delft,
C.Bountra,
O.Gileadi,
J.A.Newman.
Pandda Analysis Group Deposition To Be Published.
Page generated: Thu Aug 1 14:38:23 2024
Last articles
F in 4D85F in 4D7O
F in 4D44
F in 4D6Y
F in 4D7J
F in 4D7I
F in 4D7H
F in 4D6U
F in 4D6T
F in 4D5H