Fluorine »
PDB 5vta-5wel »
5w2e »
Fluorine in PDB 5w2e: Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876
Enzymatic activity of Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876
All present enzymatic activity of Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876:
2.7.7.48; 3.4.21.98; 3.6.1.15; 3.6.4.13;
2.7.7.48; 3.4.21.98; 3.6.1.15; 3.6.4.13;
Protein crystallography data
The structure of Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876, PDB code: 5w2e
was solved by
C.A.Lesburg,
A.Ummat,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 34.15 / 2.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 86.092, 106.418, 126.091, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.1 / 23.9 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876
(pdb code 5w2e). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 4 binding sites of Fluorine where determined in the Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876, PDB code: 5w2e:
Jump to Fluorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Fluorine where determined in the Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876, PDB code: 5w2e:
Jump to Fluorine binding site number: 1; 2; 3; 4;
Fluorine binding site 1 out of 4 in 5w2e
Go back to
Fluorine binding site 1 out
of 4 in the Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876
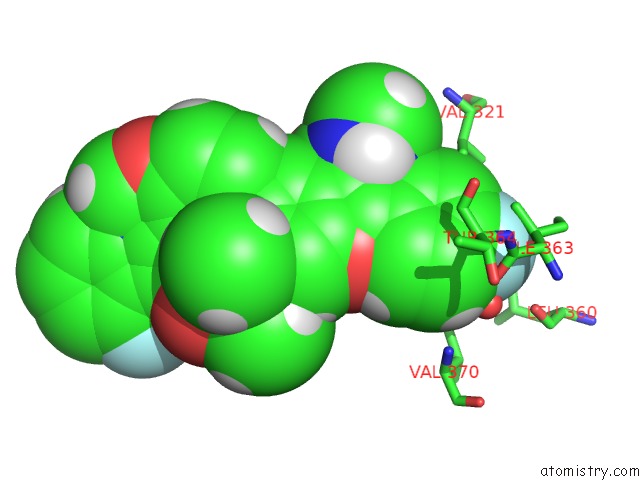
Mono view
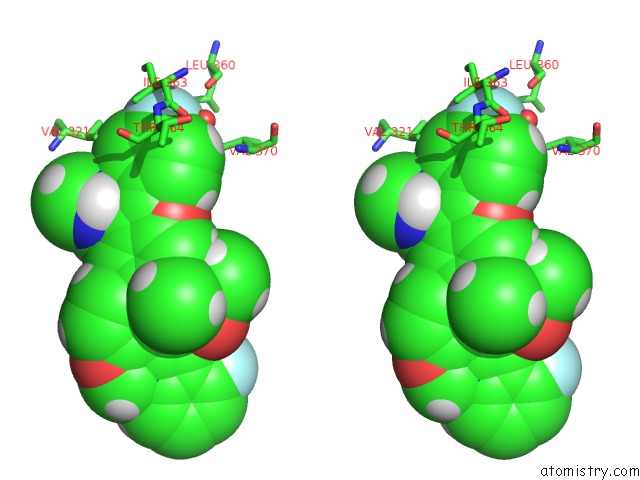
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876 within 5.0Å range:
|
Fluorine binding site 2 out of 4 in 5w2e
Go back to
Fluorine binding site 2 out
of 4 in the Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876
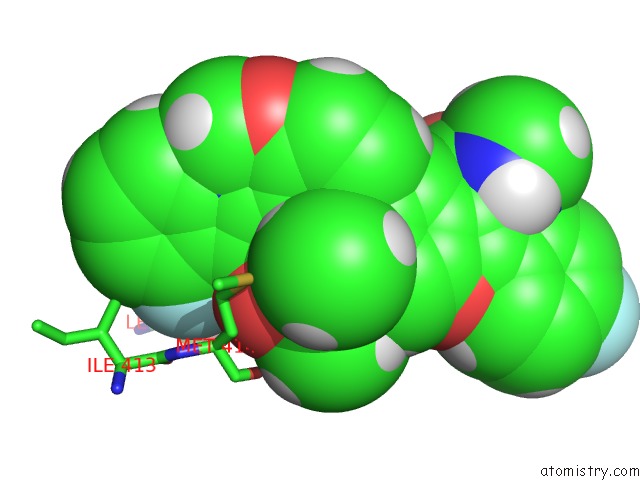
Mono view
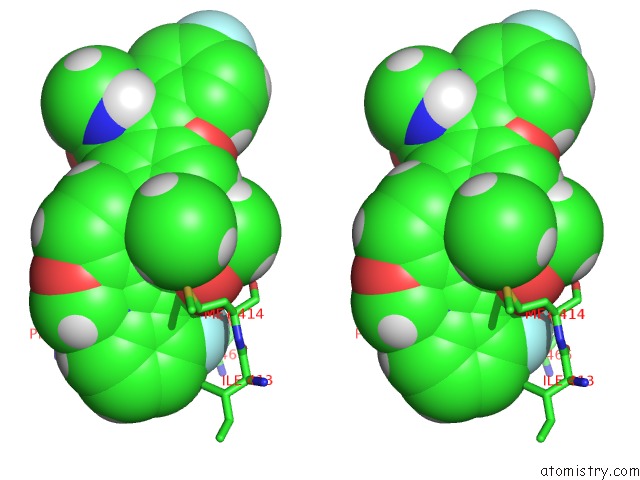
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876 within 5.0Å range:
|
Fluorine binding site 3 out of 4 in 5w2e
Go back to
Fluorine binding site 3 out
of 4 in the Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876
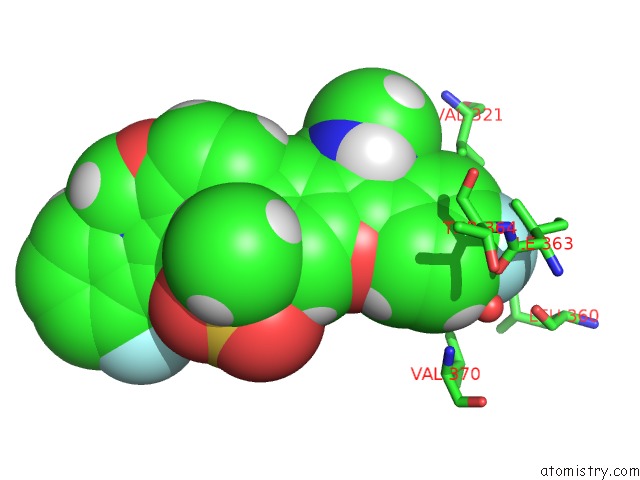
Mono view
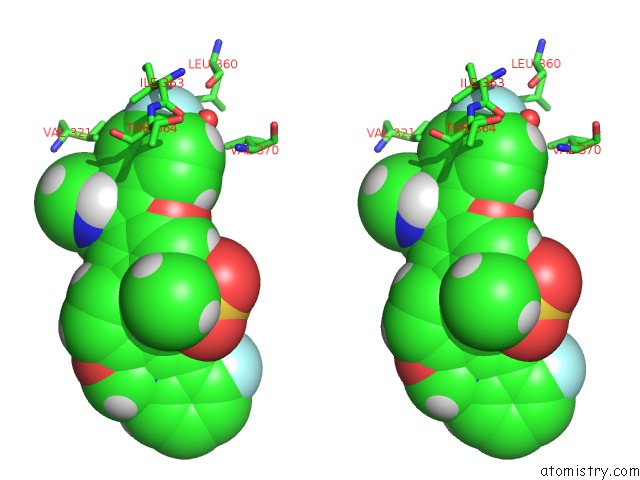
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876 within 5.0Å range:
|
Fluorine binding site 4 out of 4 in 5w2e
Go back to
Fluorine binding site 4 out
of 4 in the Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876
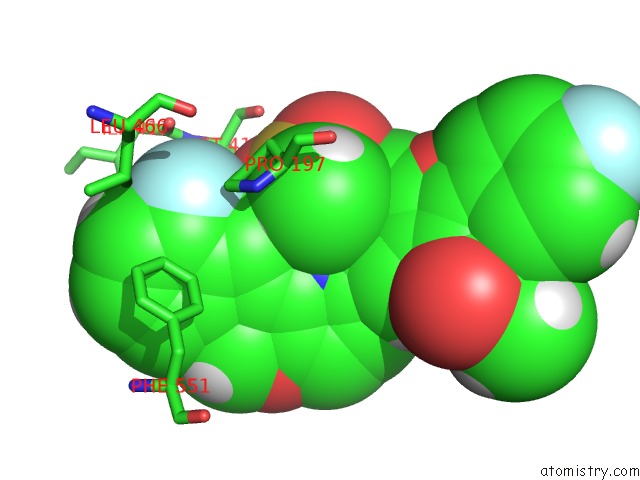
Mono view
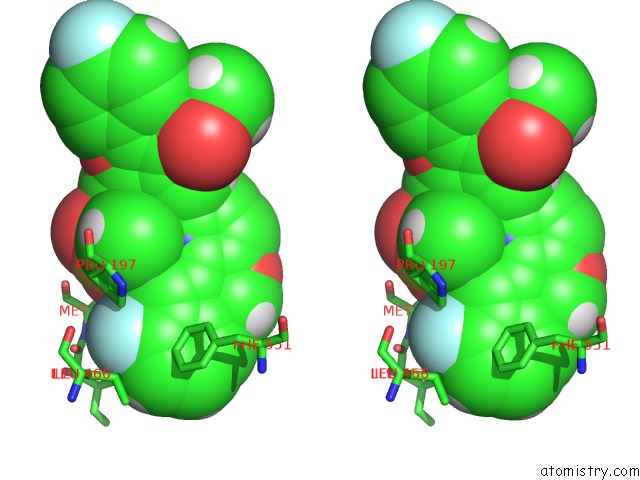
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 4 of Hcv NS5B Rna-Dependent Rna Polymerase in Complex with Non-Nucleoside Inhibitor Mk-8876 within 5.0Å range:
|
Reference:
C.C.Mccomas,
A.Palani,
W.Chang,
M.K.Holloway,
C.A.Lesburg,
P.Li,
N.Liverton,
P.T.Meinke,
D.B.Olsen,
X.Peng,
R.M.Soll,
A.Ummat,
J.Wu,
J.Wu,
N.Zorn,
S.W.Ludmerer.
Development of A New Structural Class of Broadly Acting Hcv Non-Nucleoside Inhibitors Leading to the Discovery of Mk-8876. Chemmedchem V. 12 1436 2017.
ISSN: ESSN 1860-7187
PubMed: 28741898
DOI: 10.1002/CMDC.201700228
Page generated: Thu Aug 1 16:22:21 2024
ISSN: ESSN 1860-7187
PubMed: 28741898
DOI: 10.1002/CMDC.201700228
Last articles
Cl in 5RZ4Cl in 5S2Y
Cl in 5S2W
Cl in 5S2U
Cl in 5S2N
Cl in 5RZO
Cl in 5RZG
Cl in 5RYS
Cl in 5RYN
Cl in 5RY8