Fluorine »
PDB 5xd5-5y5r »
5xmv »
Fluorine in PDB 5xmv: Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
Enzymatic activity of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
All present enzymatic activity of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362:
2.1.2.1;
2.1.2.1;
Protein crystallography data
The structure of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362, PDB code: 5xmv
was solved by
P.Chitnumsub,
A.Jaruwat,
U.Leartsakulpanich,
G.Schwertz,
F.Diederich,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 2.16 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 101.674, 58.843, 234.899, 90.00, 90.14, 90.00 |
| R / Rfree (%) | 24.2 / 30.6 |
Other elements in 5xmv:
The structure of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362 also contains other interesting chemical elements:
| Chlorine | (Cl) | 5 atoms |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
(pdb code 5xmv). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 9 binding sites of Fluorine where determined in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362, PDB code: 5xmv:
Jump to Fluorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
In total 9 binding sites of Fluorine where determined in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362, PDB code: 5xmv:
Jump to Fluorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
Fluorine binding site 1 out of 9 in 5xmv
Go back to
Fluorine binding site 1 out
of 9 in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
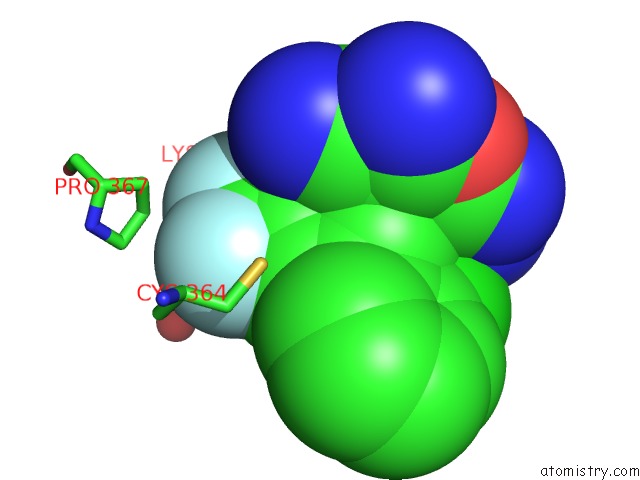
Mono view
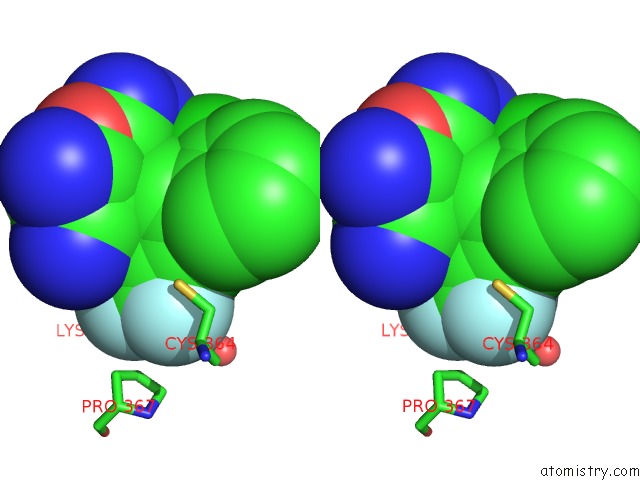
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362 within 5.0Å range:
|
Fluorine binding site 2 out of 9 in 5xmv
Go back to
Fluorine binding site 2 out
of 9 in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
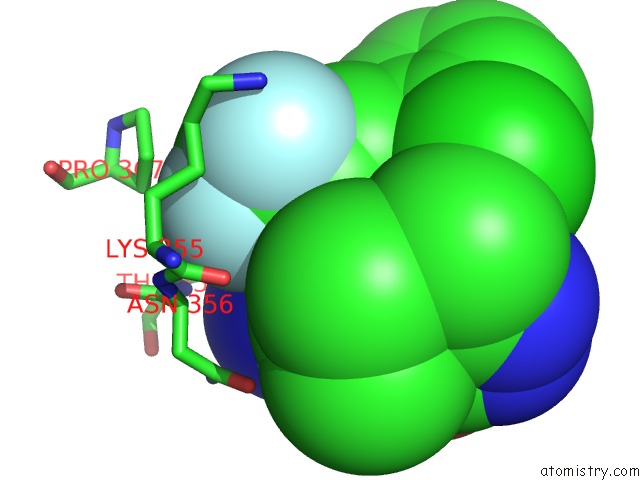
Mono view
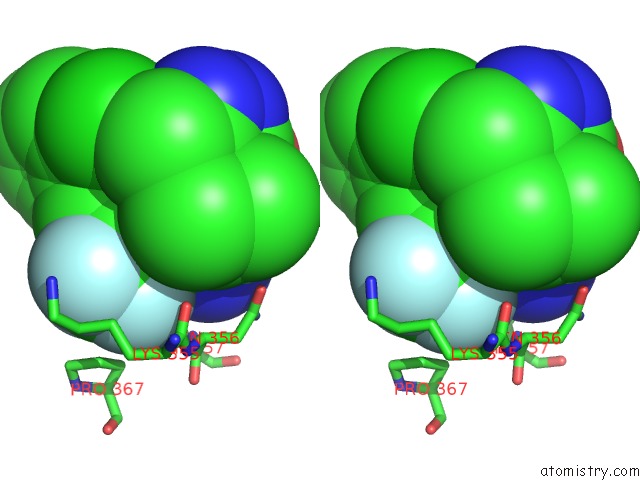
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362 within 5.0Å range:
|
Fluorine binding site 3 out of 9 in 5xmv
Go back to
Fluorine binding site 3 out
of 9 in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362 within 5.0Å range:
|
Fluorine binding site 4 out of 9 in 5xmv
Go back to
Fluorine binding site 4 out
of 9 in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 4 of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362 within 5.0Å range:
|
Fluorine binding site 5 out of 9 in 5xmv
Go back to
Fluorine binding site 5 out
of 9 in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
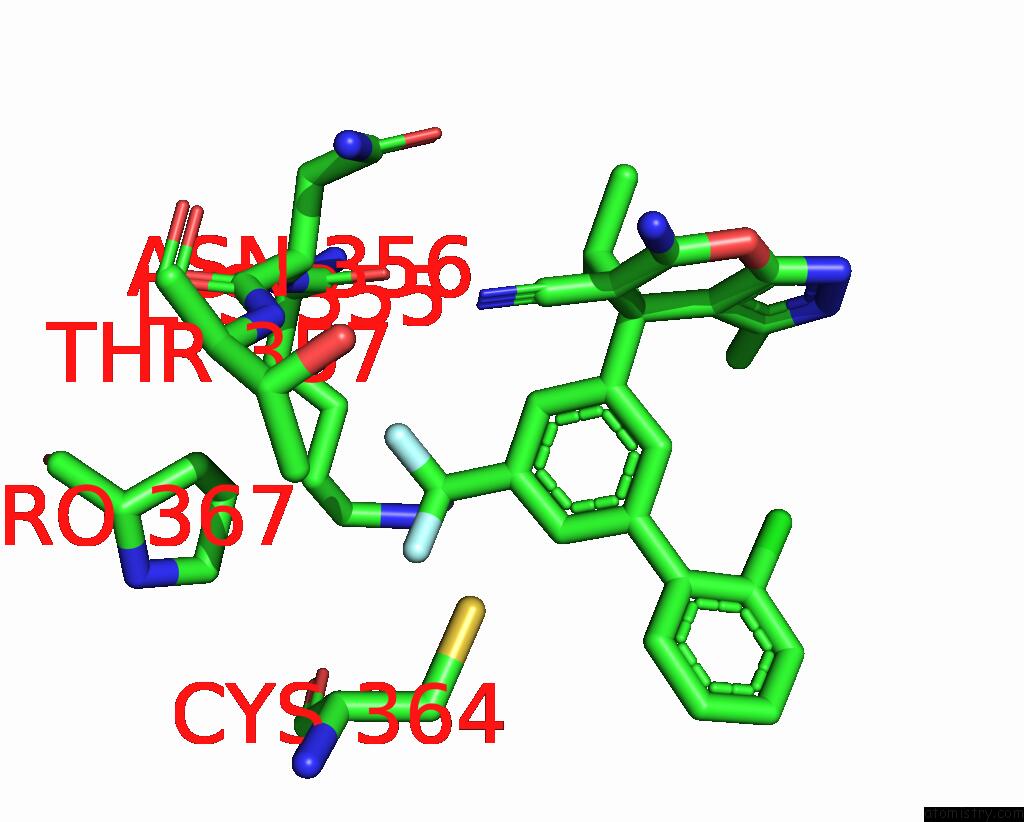
Mono view
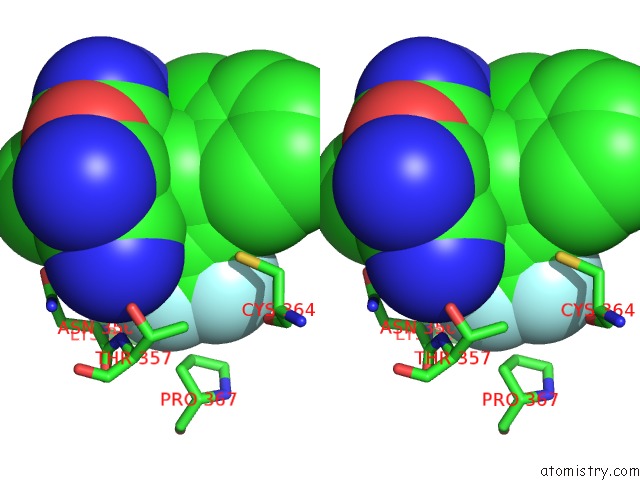
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 5 of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362 within 5.0Å range:
|
Fluorine binding site 6 out of 9 in 5xmv
Go back to
Fluorine binding site 6 out
of 9 in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
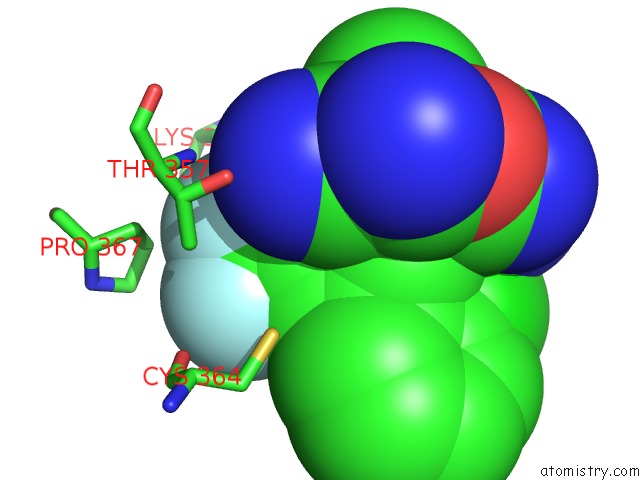
Mono view
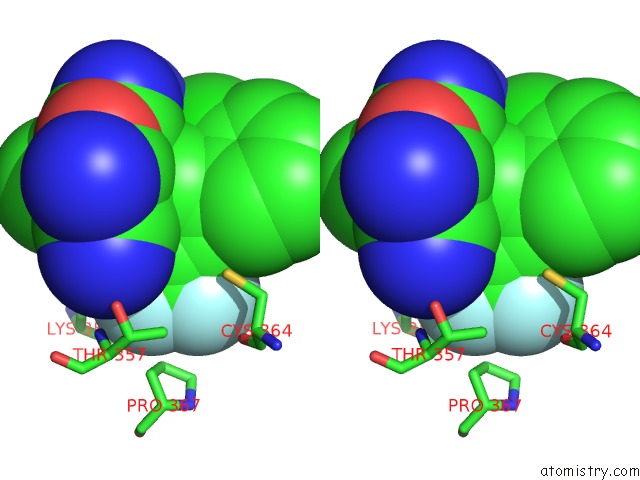
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 6 of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362 within 5.0Å range:
|
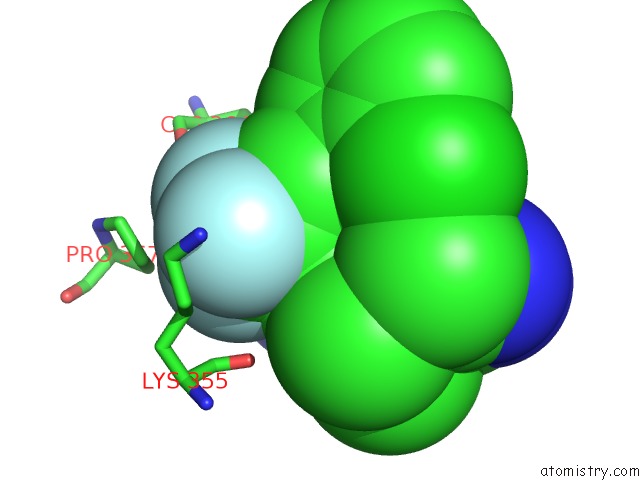
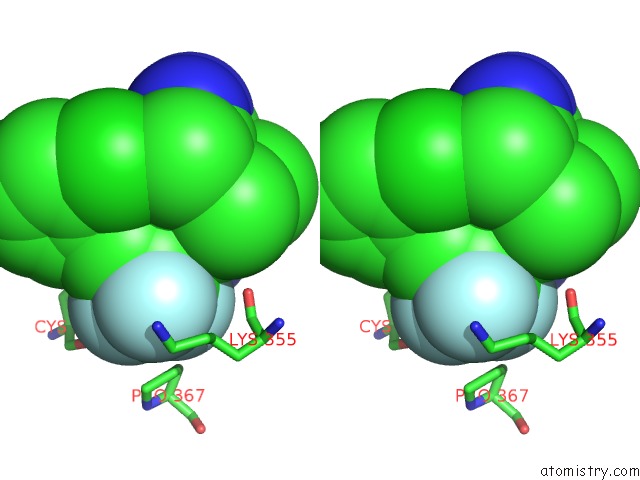
Fluorine binding site 7 out of 9 in 5xmv
Go back to
Fluorine binding site 7 out
of 9 in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 7 of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362 within 5.0Å range:
|
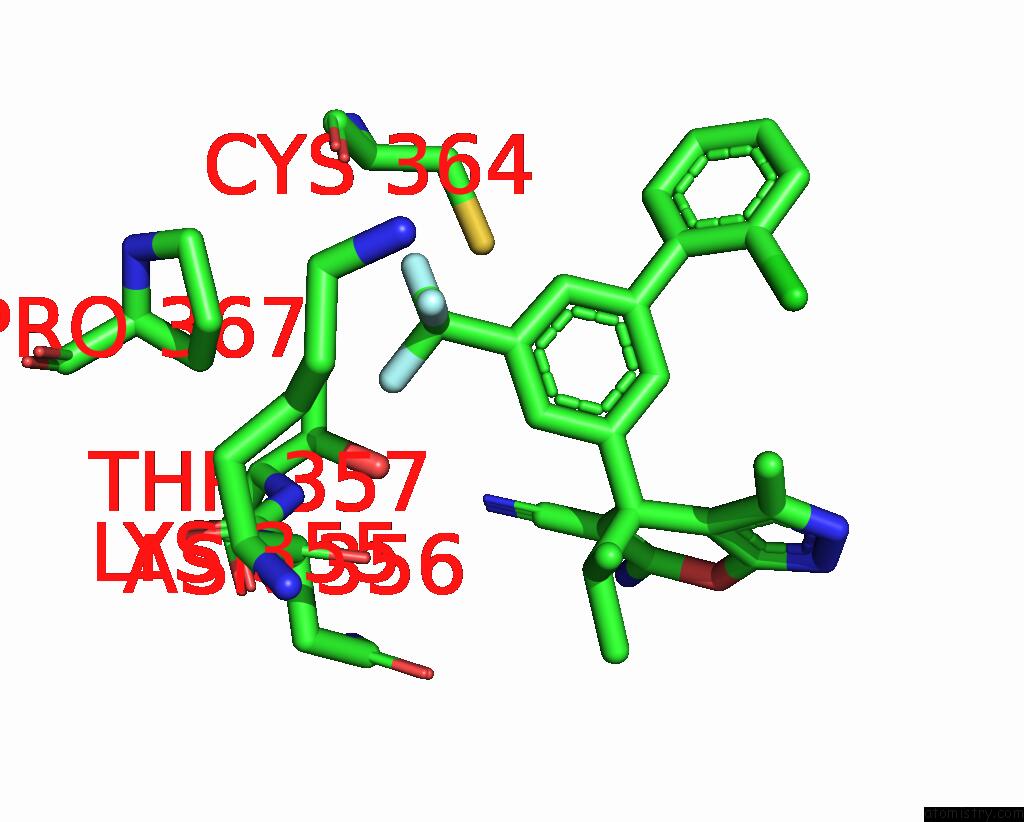
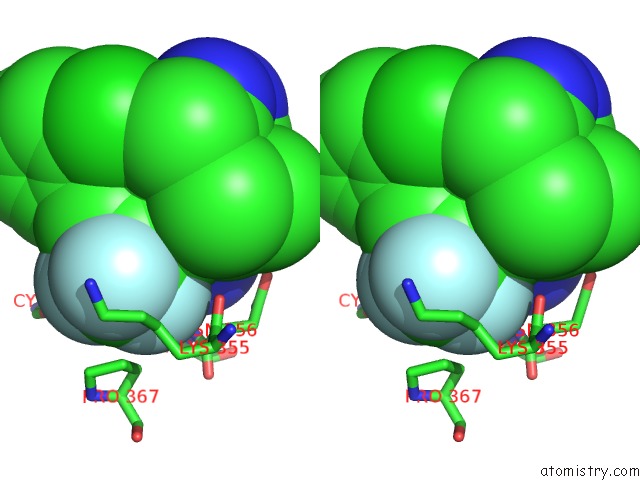
Fluorine binding site 8 out of 9 in 5xmv
Go back to
Fluorine binding site 8 out
of 9 in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 8 of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362 within 5.0Å range:
|
Fluorine binding site 9 out of 9 in 5xmv
Go back to
Fluorine binding site 9 out
of 9 in the Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362
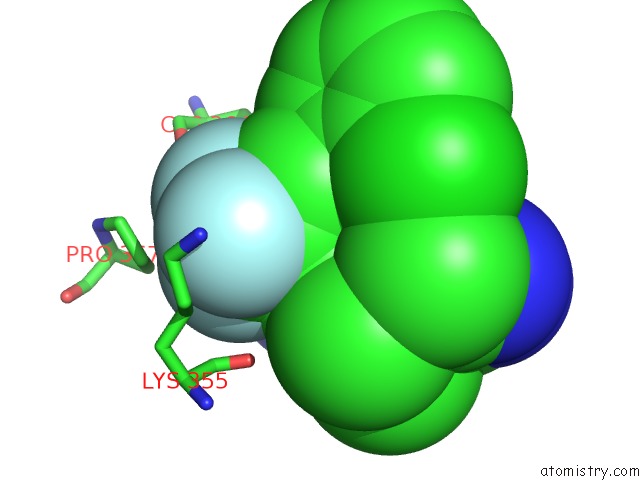
Mono view
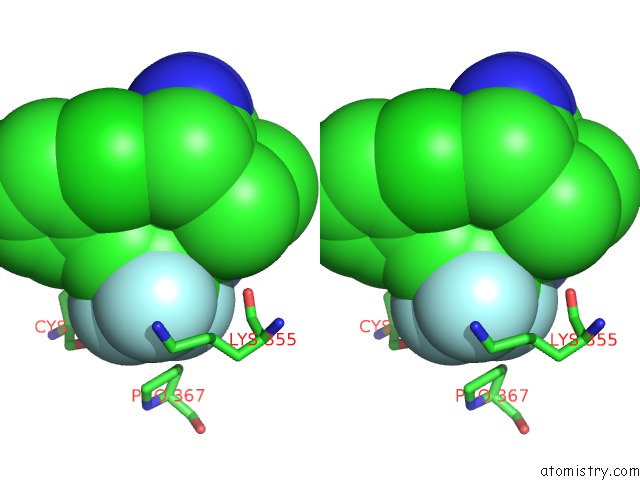
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 9 of Plasmodium Vivax Shmt Bound with Plp-Glycine and GS362 within 5.0Å range:
|
Reference:
G.Schwertz,
M.S.Frei,
M.C.Witschel,
M.Rottmann,
U.Leartsakulpanich,
P.Chitnumsub,
A.Jaruwat,
W.Ittarat,
A.Schafer,
R.A.Aponte,
N.Trapp,
K.Mark,
P.Chaiyen,
F.Diederich.
Conformational Aspects in the Design of Inhibitors For Serine Hydroxymethyltransferase (Shmt): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs Chemistry V. 23 14345 2017.
ISSN: ISSN 1521-3765
PubMed: 28967982
DOI: 10.1002/CHEM.201703244
Page generated: Thu Aug 1 17:00:25 2024
ISSN: ISSN 1521-3765
PubMed: 28967982
DOI: 10.1002/CHEM.201703244
Last articles
Ca in 5VH2Ca in 5VLK
Ca in 5VLH
Ca in 5VLA
Ca in 5VKW
Ca in 5VF4
Ca in 5VJF
Ca in 5VGC
Ca in 5VGT
Ca in 5VER