Fluorine »
PDB 5y6m-5zc5 »
5ygy »
Fluorine in PDB 5ygy: Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide
Enzymatic activity of Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide
All present enzymatic activity of Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide:
3.4.23.46;
3.4.23.46;
Protein crystallography data
The structure of Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide, PDB code: 5ygy
was solved by
K.Fuchino,
Y.Mitsuoka,
M.Masui,
N.Kurose,
S.Yoshida,
K.Komano,
T.Yamamoto,
M.Ogawa,
C.Unemura,
M.Hosono,
H.Ito,
G.Sakaguchi,
S.Ando,
S.Ohnishi,
Y.Kido,
T.Fukushima,
H.Miyajima,
S.Hiroyama,
K.Koyabu,
D.Dhuyvetter,
H.Borghys,
H.Gijsen,
Y.Yamano,
Y.Iso,
K.Kusakabe,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.30 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 102.149, 102.149, 171.125, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19.1 / 23.5 |
Other elements in 5ygy:
The structure of Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide also contains other interesting chemical elements:
| Iodine | (I) | 5 atoms |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide
(pdb code 5ygy). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide, PDB code: 5ygy:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide, PDB code: 5ygy:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 5ygy
Go back to
Fluorine binding site 1 out
of 2 in the Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide
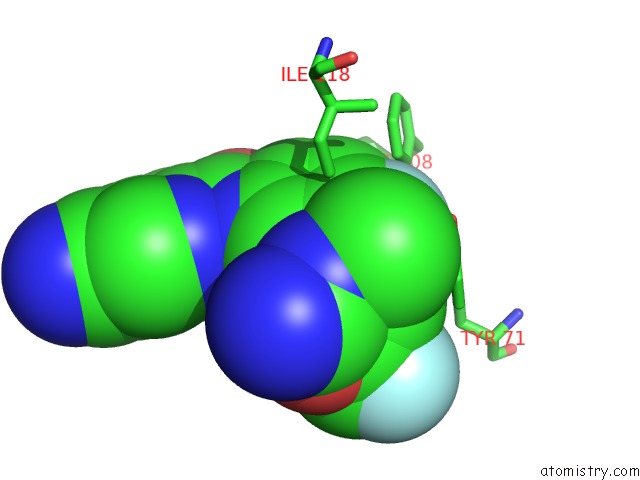
Mono view
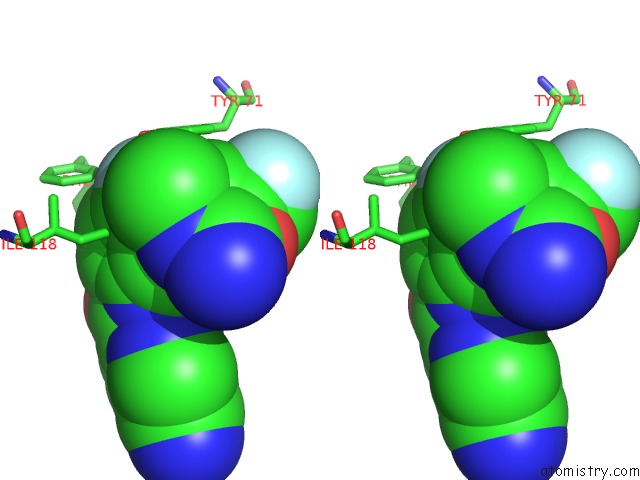
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 5ygy
Go back to
Fluorine binding site 2 out
of 2 in the Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide
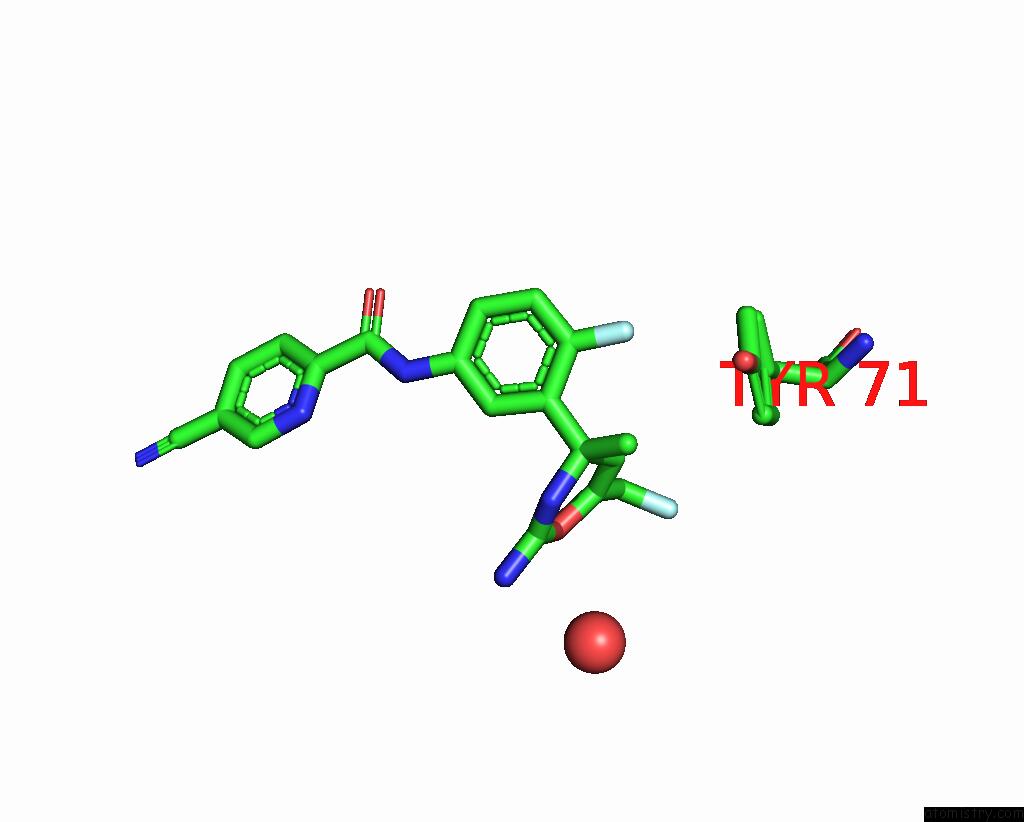
Mono view
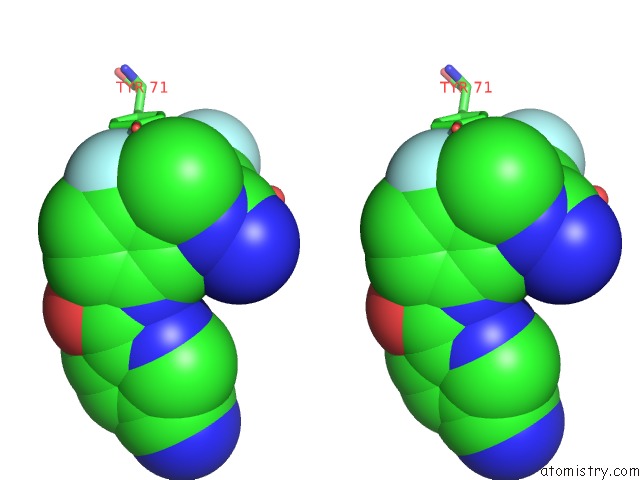
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of BACE1 in Complex with (S)-N-(3-(2-Amino-6- (Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5- Cyanopicolinamide within 5.0Å range:
|
Reference:
K.Fuchino,
Y.Mitsuoka,
M.Masui,
N.Kurose,
S.Yoshida,
K.Komano,
T.Yamamoto,
M.Ogawa,
C.Unemura,
M.Hosono,
H.Ito,
G.Sakaguchi,
S.Ando,
S.Ohnishi,
Y.Kido,
T.Fukushima,
H.Miyajima,
S.Hiroyama,
K.Koyabu,
D.Dhuyvetter,
H.Borghys,
H.J.M.Gijsen,
Y.Yamano,
Y.Iso,
K.I.Kusakabe.
Rational Design of Novel 1,3-Oxazine Based Beta-Secretase (BACE1) Inhibitors: Incorporation of A Double Bond to Reduce P-Gp Efflux Leading to Robust A Beta Reduction in the Brain J. Med. Chem. V. 61 5122 2018.
ISSN: ISSN 1520-4804
PubMed: 29733614
DOI: 10.1021/ACS.JMEDCHEM.8B00002
Page generated: Tue Jul 15 09:30:15 2025
ISSN: ISSN 1520-4804
PubMed: 29733614
DOI: 10.1021/ACS.JMEDCHEM.8B00002
Last articles
F in 6RV3F in 6RS5
F in 6RSZ
F in 6RSU
F in 6RST
F in 6RSR
F in 6RRI
F in 6RSE
F in 6RSH
F in 6RSB