Fluorine »
PDB 6c0u-6cr2 »
6cbh »
Fluorine in PDB 6cbh: Macrophage Migration Inhibitory Factor in Complex with A Pyrazole Inhibitor (8M)
Enzymatic activity of Macrophage Migration Inhibitory Factor in Complex with A Pyrazole Inhibitor (8M)
All present enzymatic activity of Macrophage Migration Inhibitory Factor in Complex with A Pyrazole Inhibitor (8M):
5.3.2.1; 5.3.3.12;
5.3.2.1; 5.3.3.12;
Protein crystallography data
The structure of Macrophage Migration Inhibitory Factor in Complex with A Pyrazole Inhibitor (8M), PDB code: 6cbh
was solved by
M.J.Robertson,
S.G.Krimmer,
W.L.Jorgensen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 54.20 / 2.00 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.149, 68.460, 88.699, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.3 / 21.4 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Macrophage Migration Inhibitory Factor in Complex with A Pyrazole Inhibitor (8M)
(pdb code 6cbh). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total only one binding site of Fluorine was determined in the Macrophage Migration Inhibitory Factor in Complex with A Pyrazole Inhibitor (8M), PDB code: 6cbh:
In total only one binding site of Fluorine was determined in the Macrophage Migration Inhibitory Factor in Complex with A Pyrazole Inhibitor (8M), PDB code: 6cbh:
Fluorine binding site 1 out of 1 in 6cbh
Go back to
Fluorine binding site 1 out
of 1 in the Macrophage Migration Inhibitory Factor in Complex with A Pyrazole Inhibitor (8M)
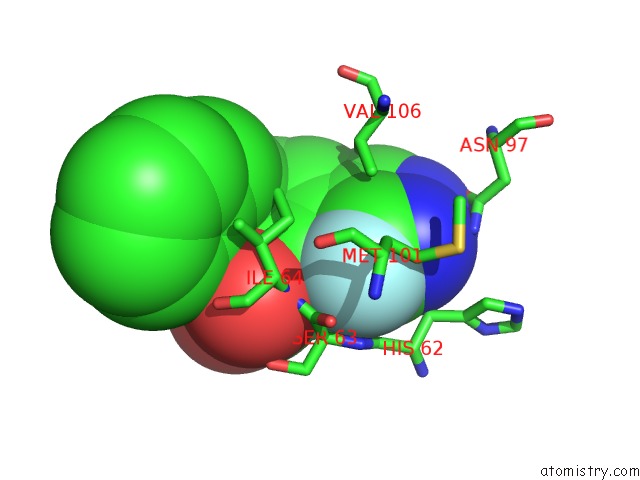
Mono view
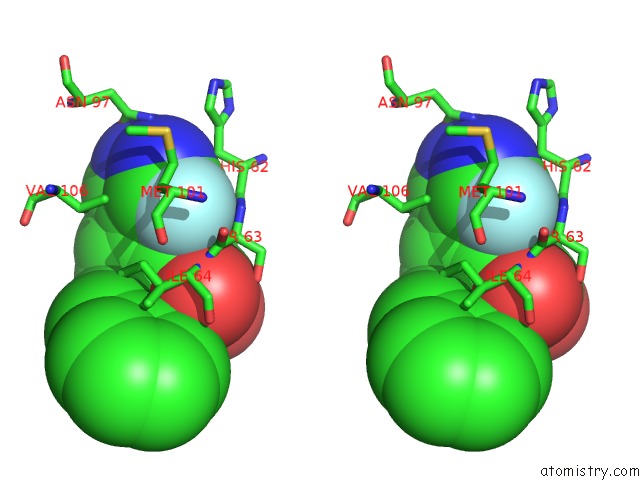
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Macrophage Migration Inhibitory Factor in Complex with A Pyrazole Inhibitor (8M) within 5.0Å range:
|
Reference:
V.Trivedi-Parmar,
M.J.Robertson,
J.A.Cisneros,
S.G.Krimmer,
W.L.Jorgensen.
Optimization of Pyrazoles As Phenol Surrogates to Yield Potent Inhibitors of Macrophage Migration Inhibitory Factor. Chemmedchem V. 13 1092 2018.
ISSN: ESSN 1860-7187
PubMed: 29575754
DOI: 10.1002/CMDC.201800158
Page generated: Tue Jul 15 10:21:22 2025
ISSN: ESSN 1860-7187
PubMed: 29575754
DOI: 10.1002/CMDC.201800158
Last articles
F in 6TQSF in 6TTZ
F in 6TUF
F in 6TUC
F in 6TTI
F in 6TU9
F in 6TRX
F in 6TQ6
F in 6TQ9
F in 6TR7