Fluorine »
PDB 6dmg-6e69 »
6dnp »
Fluorine in PDB 6dnp: Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid
Enzymatic activity of Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid
All present enzymatic activity of Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid:
2.3.3.9;
2.3.3.9;
Protein crystallography data
The structure of Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid, PDB code: 6dnp
was solved by
I.V.Krieger,
J.C.Sacchettini,
Tb Structural Genomics Consortium(Tbsgc),
Mycobacterium Tuberculosis Structural Proteomics Project(Xmtb),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.62 / 1.71 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 79.737, 79.737, 227.288, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.7 / 25.1 |
Other elements in 6dnp:
The structure of Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid
(pdb code 6dnp). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid, PDB code: 6dnp:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid, PDB code: 6dnp:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 6dnp
Go back to
Fluorine binding site 1 out
of 2 in the Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid
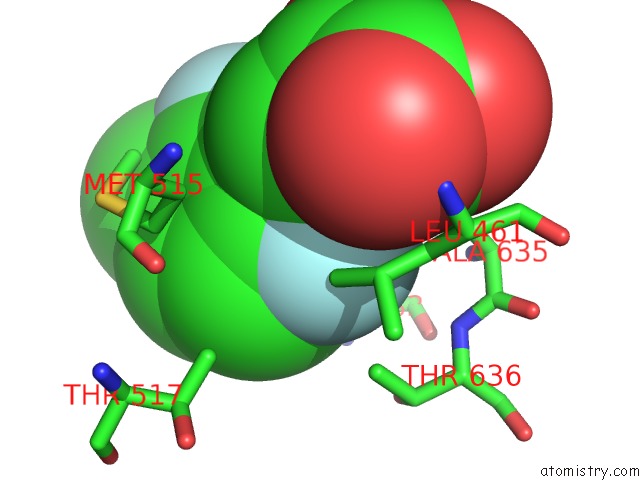
Mono view
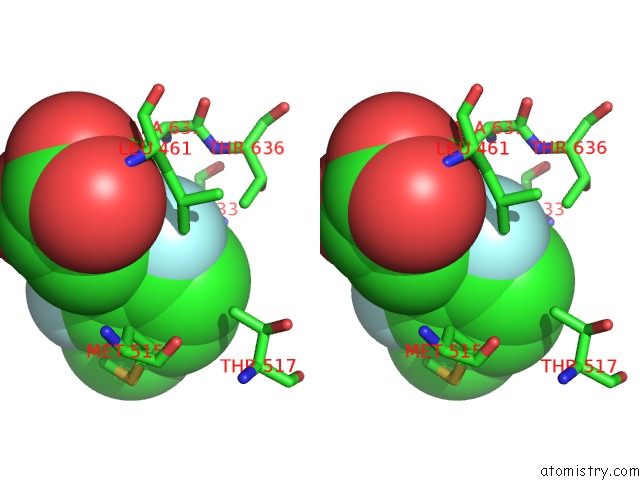
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 6dnp
Go back to
Fluorine binding site 2 out
of 2 in the Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid
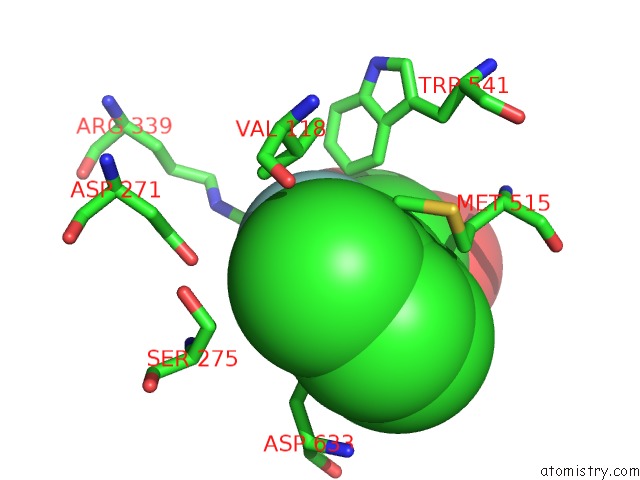
Mono view
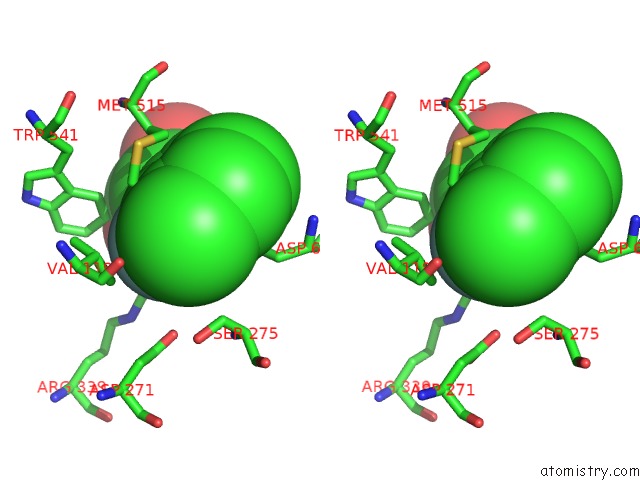
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of Mycobacterium Tuberculosis Malate Synthase in Complex with 2-F-3-Methyl-6-F-Phenyldiketoacid within 5.0Å range:
|
Reference:
J.F.Ellenbarger,
I.V.Krieger,
H.L.Huang,
S.Gomez-Coca,
T.R.Ioerger,
J.C.Sacchettini,
S.E.Wheeler,
K.R.Dunbar.
Anion-Pi Interactions in Computer-Aided Drug Design: Modeling the Inhibition of Malate Synthase By Phenyl-Diketo Acids. J Chem Inf Model V. 58 2085 2018.
ISSN: ESSN 1549-960X
PubMed: 30137983
DOI: 10.1021/ACS.JCIM.8B00417
Page generated: Tue Jul 15 10:50:18 2025
ISSN: ESSN 1549-960X
PubMed: 30137983
DOI: 10.1021/ACS.JCIM.8B00417
Last articles
F in 6WQQF in 6WQE
F in 6WQ7
F in 6WPP
F in 6WPH
F in 6WOI
F in 6WOB
F in 6WOA
F in 6WO8
F in 6WNY