Fluorine »
PDB 6jsg-6kk1 »
6jyl »
Fluorine in PDB 6jyl: The Crosslinked Complex of Iswi-Nucleosome in the Adp.Bef-Bound State
Other elements in 6jyl:
The structure of The Crosslinked Complex of Iswi-Nucleosome in the Adp.Bef-Bound State also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the The Crosslinked Complex of Iswi-Nucleosome in the Adp.Bef-Bound State
(pdb code 6jyl). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the The Crosslinked Complex of Iswi-Nucleosome in the Adp.Bef-Bound State, PDB code: 6jyl:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the The Crosslinked Complex of Iswi-Nucleosome in the Adp.Bef-Bound State, PDB code: 6jyl:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 6jyl
Go back to
Fluorine binding site 1 out
of 3 in the The Crosslinked Complex of Iswi-Nucleosome in the Adp.Bef-Bound State

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of The Crosslinked Complex of Iswi-Nucleosome in the Adp.Bef-Bound State within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 6jyl
Go back to
Fluorine binding site 2 out
of 3 in the The Crosslinked Complex of Iswi-Nucleosome in the Adp.Bef-Bound State

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of The Crosslinked Complex of Iswi-Nucleosome in the Adp.Bef-Bound State within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 6jyl
Go back to
Fluorine binding site 3 out
of 3 in the The Crosslinked Complex of Iswi-Nucleosome in the Adp.Bef-Bound State
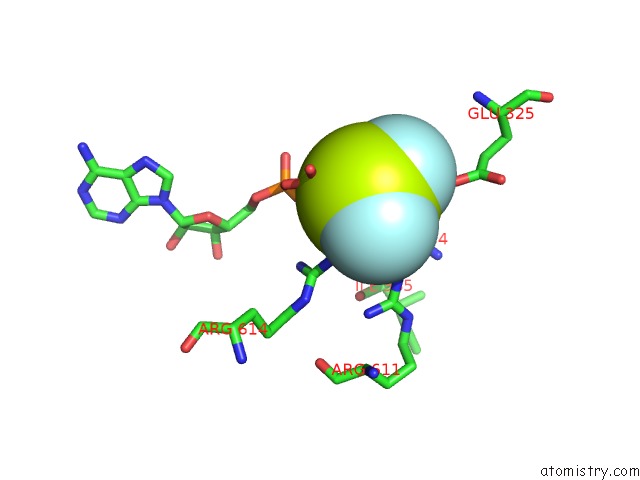
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of The Crosslinked Complex of Iswi-Nucleosome in the Adp.Bef-Bound State within 5.0Å range:
|
Reference:
L.Yan,
H.Wu,
X.Li,
N.Gao,
Z.Chen.
Structures of the Iswi-Nucleosome Complex Reveal A Conserved Mechanism of Chromatin Remodeling. Nat.Struct.Mol.Biol. V. 26 258 2019.
ISSN: ESSN 1545-9985
PubMed: 30872815
DOI: 10.1038/S41594-019-0199-9
Page generated: Tue Jul 15 12:45:12 2025
ISSN: ESSN 1545-9985
PubMed: 30872815
DOI: 10.1038/S41594-019-0199-9
Last articles
F in 7AX4F in 7AW3
F in 7AWR
F in 7AW1
F in 7AVS
F in 7AVU
F in 7AW0
F in 7AVZ
F in 7AVX
F in 7AVV