Fluorine »
PDB 6ltk-6mh7 »
6mcs »
Fluorine in PDB 6mcs: X-Ray Crystal Structure of Wild Type Hiv-1 Protease in Complex with Grl-003
Protein crystallography data
The structure of X-Ray Crystal Structure of Wild Type Hiv-1 Protease in Complex with Grl-003, PDB code: 6mcs
was solved by
H.Bulut,
H.Hayashi,
S.I.Hattori,
M.Aoki,
D.Das,
A.K.Ghosh,
H.Mitsuya,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 32.92 / 1.52 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 62.512, 62.512, 82.946, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20.6 / 24.2 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the X-Ray Crystal Structure of Wild Type Hiv-1 Protease in Complex with Grl-003
(pdb code 6mcs). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total only one binding site of Fluorine was determined in the X-Ray Crystal Structure of Wild Type Hiv-1 Protease in Complex with Grl-003, PDB code: 6mcs:
In total only one binding site of Fluorine was determined in the X-Ray Crystal Structure of Wild Type Hiv-1 Protease in Complex with Grl-003, PDB code: 6mcs:
Fluorine binding site 1 out of 1 in 6mcs
Go back to
Fluorine binding site 1 out
of 1 in the X-Ray Crystal Structure of Wild Type Hiv-1 Protease in Complex with Grl-003
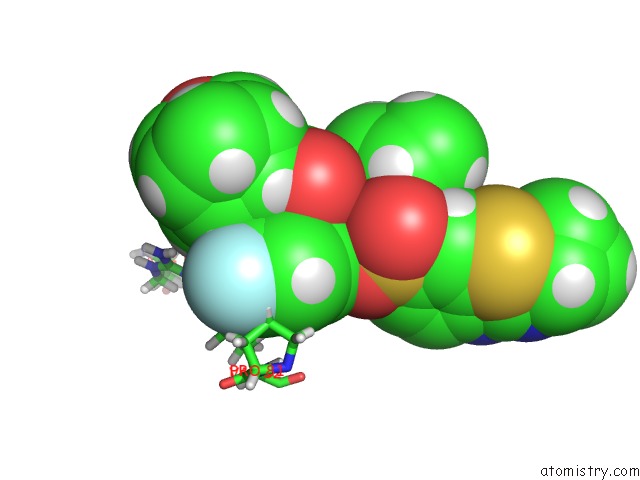
Mono view
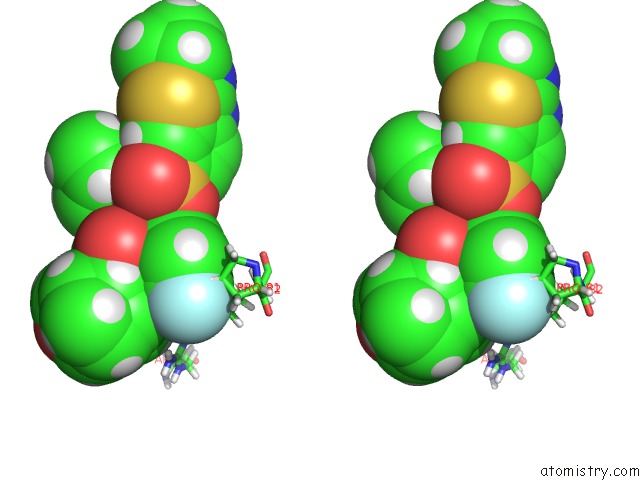
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of X-Ray Crystal Structure of Wild Type Hiv-1 Protease in Complex with Grl-003 within 5.0Å range:
|
Reference:
S.I.Hattori,
H.Hayashi,
H.Bulut,
K.V.Rao,
P.R.Nyalapatla,
K.Hasegawa,
M.Aoki,
A.K.Ghosh,
H.Mitsuya.
Halogen Bond Interactions of Novel Hiv-1 Protease Inhibitors (Pi) (Grl-001-15 and Grl-003-15) with the Flap of Protease Are Critical For Their Potent Activity Against Wild-Type Hiv-1 and Multi-Pi-Resistant Variants. Antimicrob.Agents Chemother. V. 63 2019.
ISSN: ESSN 1098-6596
PubMed: 30962341
DOI: 10.1128/AAC.02635-18
Page generated: Thu Aug 1 22:15:55 2024
ISSN: ESSN 1098-6596
PubMed: 30962341
DOI: 10.1128/AAC.02635-18
Last articles
Ca in 5NG1Ca in 5NFZ
Ca in 5NF3
Ca in 5NF2
Ca in 5NF0
Ca in 5NEY
Ca in 5NES
Ca in 5NEU
Ca in 5NET
Ca in 5NER