Fluorine »
PDB 6tvl-6ugr »
6u36 »
Fluorine in PDB 6u36: PCSK9 in Complex with A Fab and Compound 14
Protein crystallography data
The structure of PCSK9 in Complex with A Fab and Compound 14, PDB code: 6u36
was solved by
J.Lu,
S.Soisson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.67 / 2.70 |
| Space group | P 65 |
| Cell size a, b, c (Å), α, β, γ (°) | 155.605, 155.605, 152.451, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 24 / 25.1 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the PCSK9 in Complex with A Fab and Compound 14
(pdb code 6u36). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total only one binding site of Fluorine was determined in the PCSK9 in Complex with A Fab and Compound 14, PDB code: 6u36:
In total only one binding site of Fluorine was determined in the PCSK9 in Complex with A Fab and Compound 14, PDB code: 6u36:
Fluorine binding site 1 out of 1 in 6u36
Go back to
Fluorine binding site 1 out
of 1 in the PCSK9 in Complex with A Fab and Compound 14
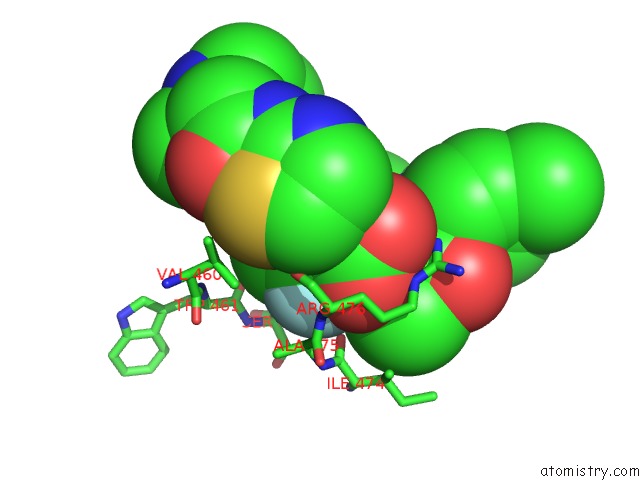
Mono view
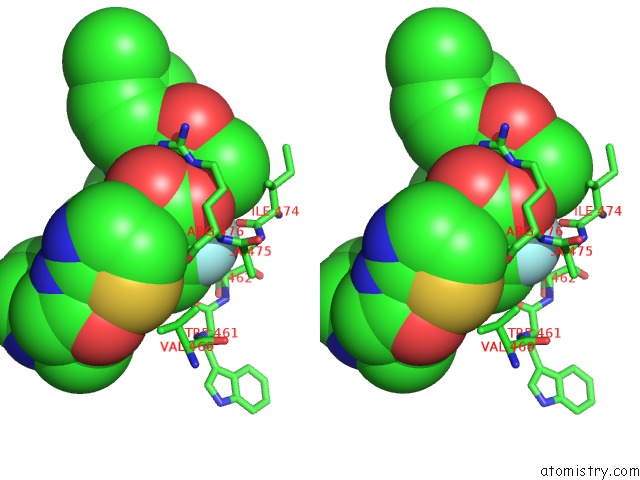
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of PCSK9 in Complex with A Fab and Compound 14 within 5.0Å range:
|
Reference:
W.L.Petrilli,
G.C.Adam,
R.S.Erdmann,
P.Abeywickrema,
V.Agnani,
X.Ai,
J.Baysarowich,
N.Byrne,
J.P.Caldwell,
W.Chang,
E.Dinunzio,
Z.Feng,
R.Ford,
S.Ha,
Y.Huang,
B.Hubbard,
J.M.Johnston,
M.Kavana,
J.M.Lisnock,
R.Liang,
J.Lu,
Z.Lu,
J.Meng,
P.Orth,
O.Palyha,
G.Parthasarathy,
S.P.Salowe,
S.Sharma,
J.Shipman,
S.M.Soisson,
A.M.Strack,
H.Youm,
K.Zhao,
D.L.Zink,
H.Zokian,
G.H.Addona,
K.Akinsanya,
J.R.Tata,
Y.Xiong,
J.E.Imbriglio.
From Screening to Targeted Degradation: Strategies For the Discovery and Optimization of Small Molecule Ligands For PCSK9. Cell Chem Biol 2019.
ISSN: ESSN 2451-9456
PubMed: 31653597
DOI: 10.1016/J.CHEMBIOL.2019.10.002
Page generated: Tue Jul 15 16:08:07 2025
ISSN: ESSN 2451-9456
PubMed: 31653597
DOI: 10.1016/J.CHEMBIOL.2019.10.002
Last articles
F in 7M01F in 7M03
F in 7M02
F in 7LZU
F in 7LZW
F in 7LY8
F in 7LWG
F in 7LZV
F in 7LZF
F in 7LZD