Fluorine »
PDB 7adu-7ayj »
7as2 »
Fluorine in PDB 7as2: Influenza A PB2 (M431 Mutation) in Complex with Vx-787
Protein crystallography data
The structure of Influenza A PB2 (M431 Mutation) in Complex with Vx-787, PDB code: 7as2
was solved by
K.Radilova,
J.Brynda,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.34 / 1.75 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 29.19, 37.12, 38.45, 71.85, 75.39, 75.92 |
| R / Rfree (%) | 16 / 20.5 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Influenza A PB2 (M431 Mutation) in Complex with Vx-787
(pdb code 7as2). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Influenza A PB2 (M431 Mutation) in Complex with Vx-787, PDB code: 7as2:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Influenza A PB2 (M431 Mutation) in Complex with Vx-787, PDB code: 7as2:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 7as2
Go back to
Fluorine binding site 1 out
of 2 in the Influenza A PB2 (M431 Mutation) in Complex with Vx-787
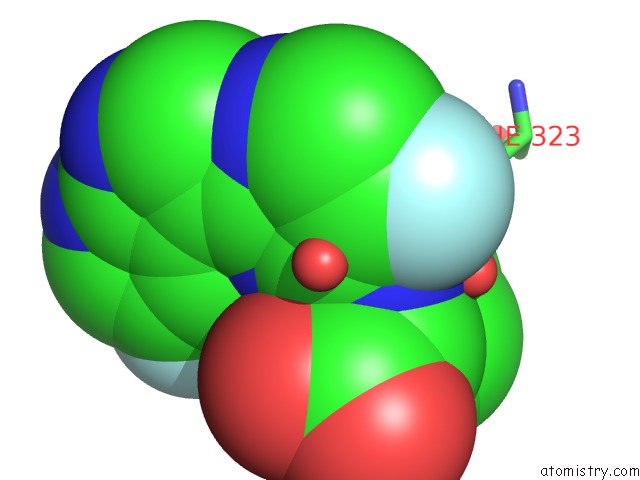
Mono view
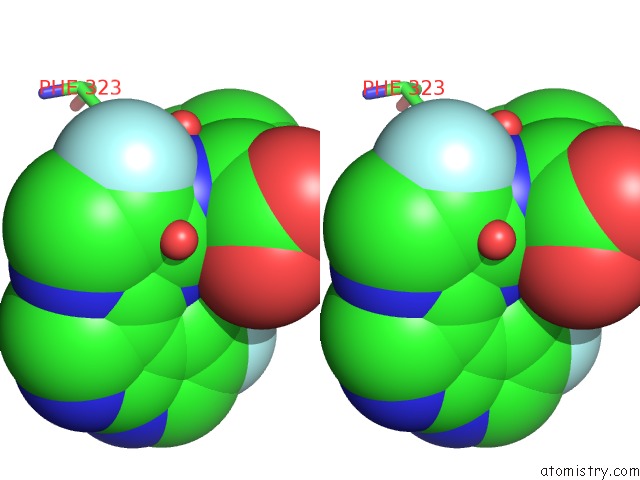
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Influenza A PB2 (M431 Mutation) in Complex with Vx-787 within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 7as2
Go back to
Fluorine binding site 2 out
of 2 in the Influenza A PB2 (M431 Mutation) in Complex with Vx-787
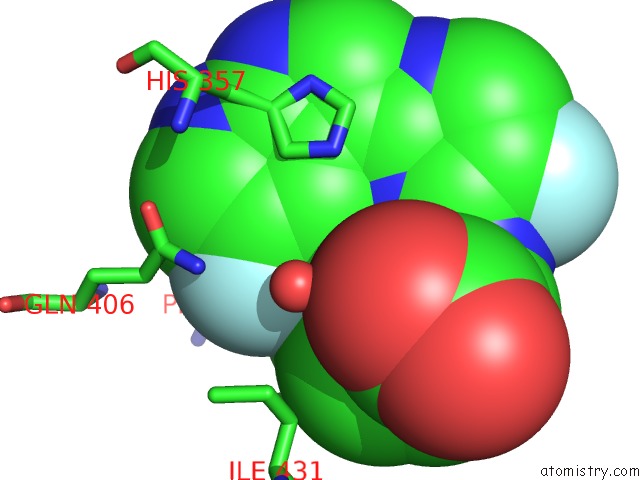
Mono view
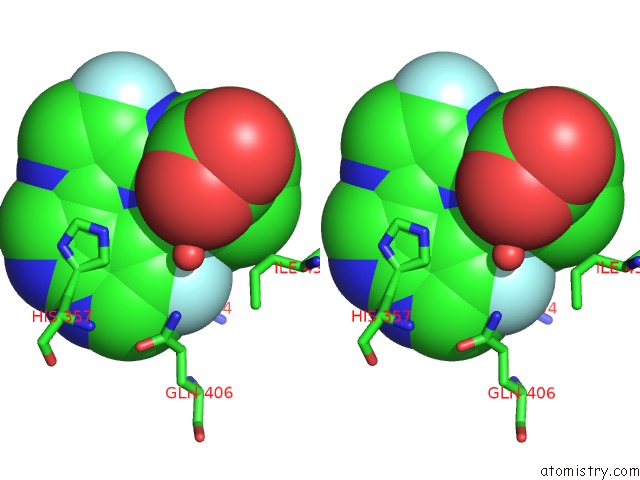
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Influenza A PB2 (M431 Mutation) in Complex with Vx-787 within 5.0Å range:
|
Reference:
J.Gregor,
K.Radilova,
J.Brynda,
J.Fanfrlik,
J.Konvalinka,
M.Kozisek.
Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (Vx-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase Molecules 2021.
ISSN: ESSN 1420-3049
DOI: 10.3390/MOLECULES26041007
Page generated: Fri Aug 2 05:43:19 2024
ISSN: ESSN 1420-3049
DOI: 10.3390/MOLECULES26041007
Last articles
Ca in 5S4UCa in 5S4T
Ca in 5S4R
Ca in 5S4S
Ca in 5S4P
Ca in 5S4Q
Ca in 5S4O
Ca in 5S4N
Ca in 5S4M
Ca in 5S4L