Fluorine »
PDB 8gky-8hej »
8gua »
Fluorine in PDB 8gua: Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719
Enzymatic activity of Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719
All present enzymatic activity of Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719:
2.7.1.137; 2.7.1.153; 2.7.11.1;
2.7.1.137; 2.7.1.153; 2.7.11.1;
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719
(pdb code 8gua). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719, PDB code: 8gua:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719, PDB code: 8gua:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 8gua
Go back to
Fluorine binding site 1 out
of 3 in the Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719

Mono view
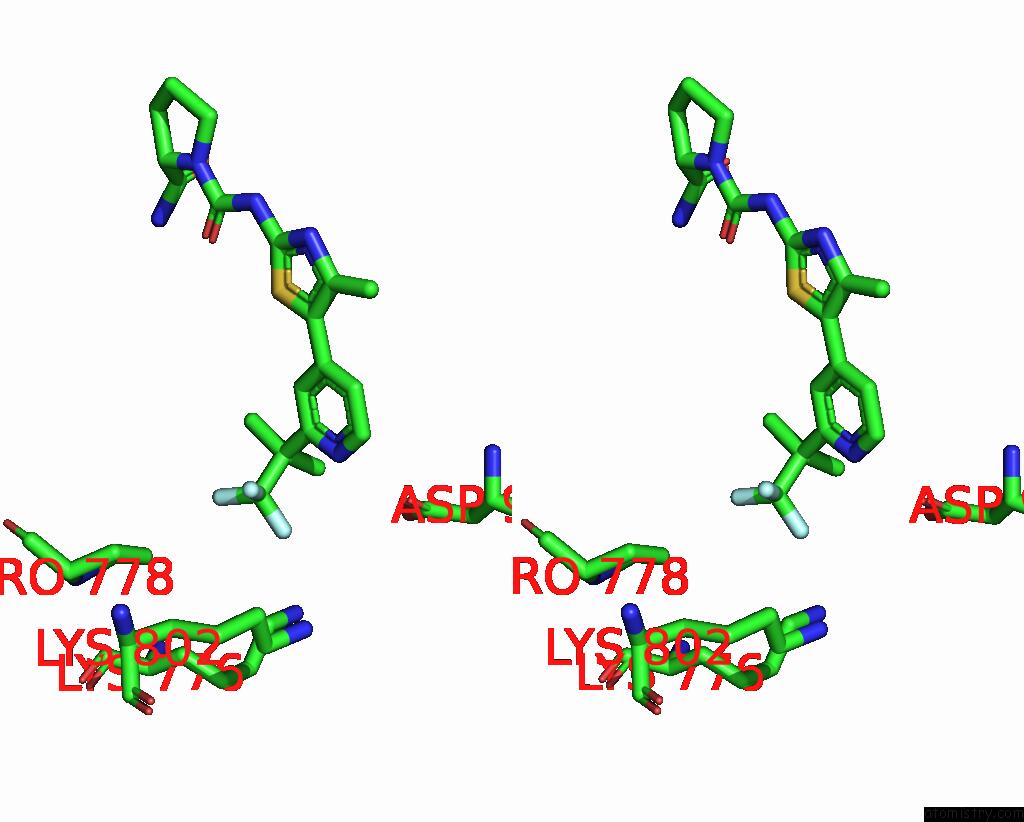
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719 within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 8gua
Go back to
Fluorine binding site 2 out
of 3 in the Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719
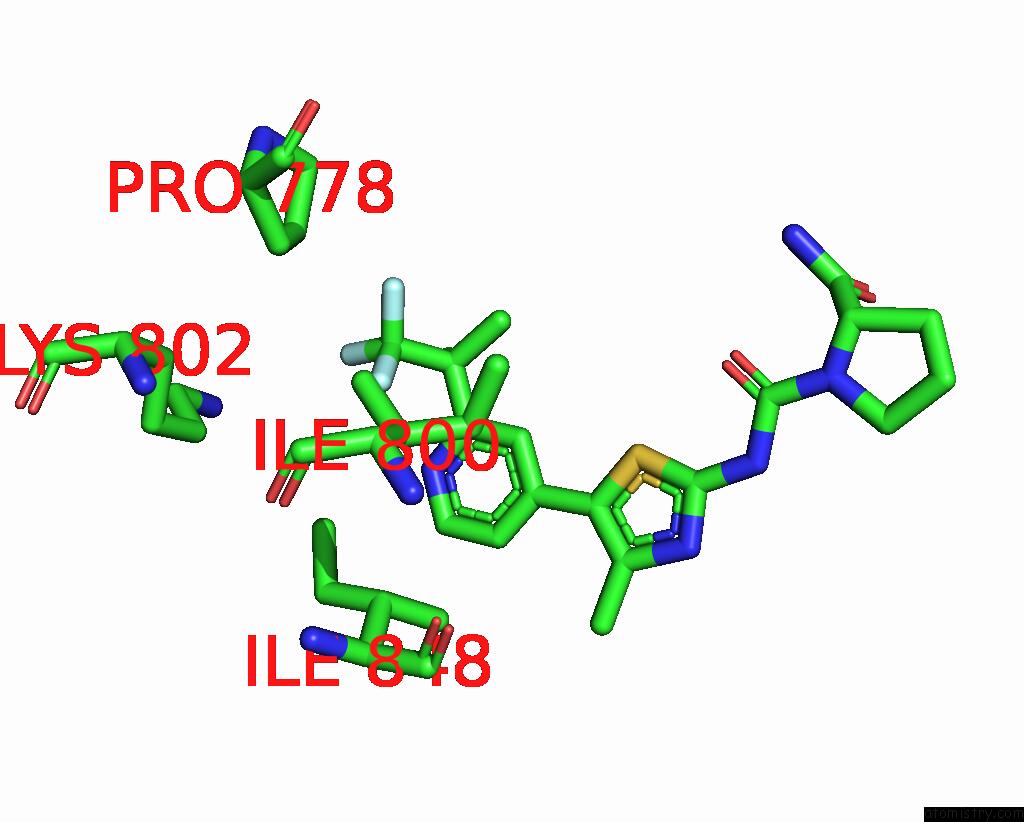
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719 within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 8gua
Go back to
Fluorine binding site 3 out
of 3 in the Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719
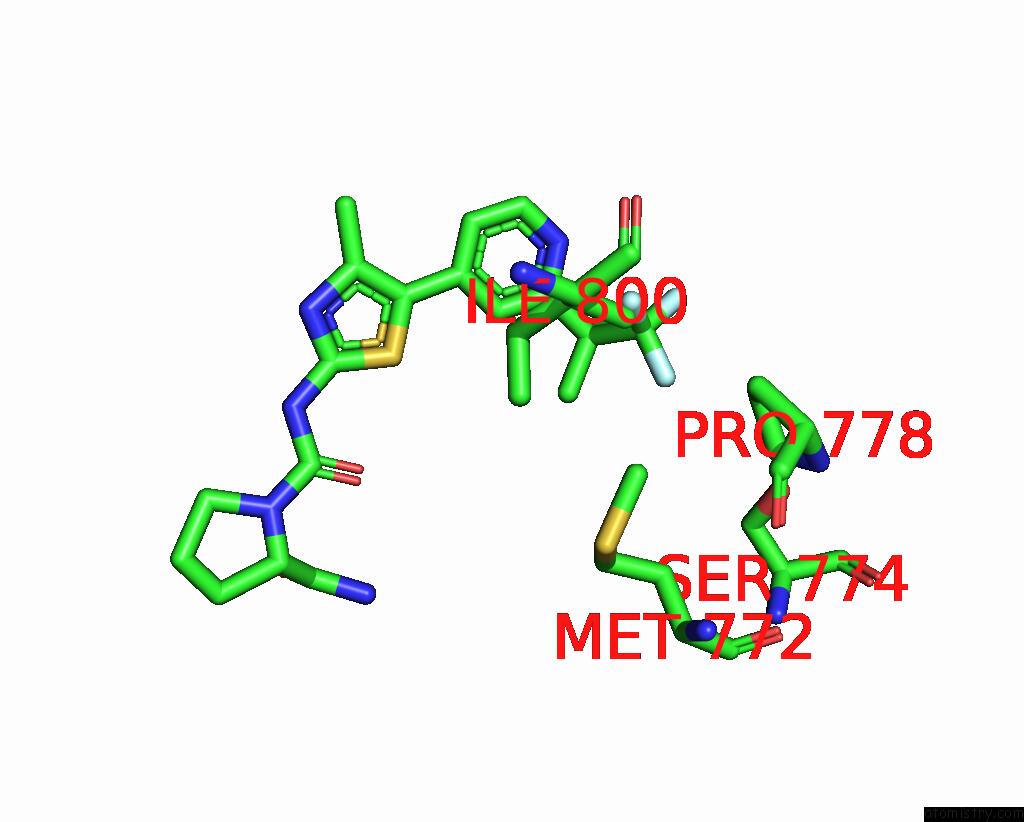
Mono view
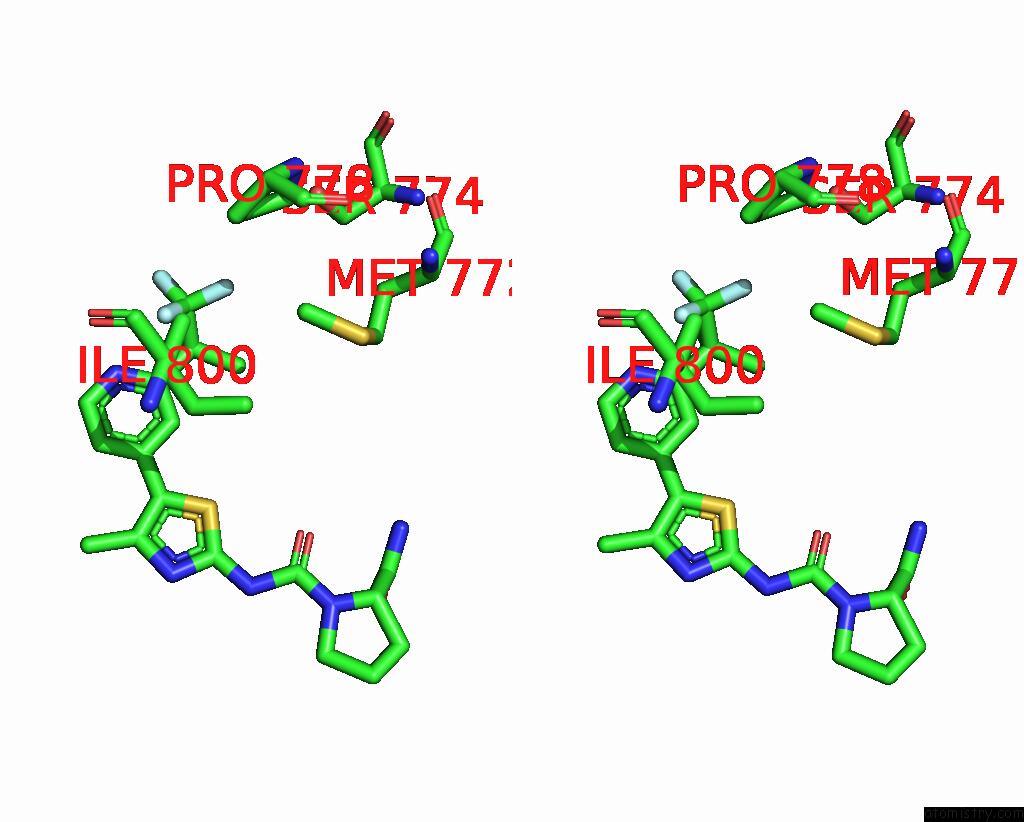
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Cryo-Em Structure of Cancer-Specific PI3KALPHA Mutant E542K in Complex with Byl-719 within 5.0Å range:
|
Reference:
X.Liu,
Q.Zhou,
J.R.Hart,
Y.Xu,
S.Yang,
D.Yang,
P.K.Vogt,
M.W.Wang.
Cryo-Em Structures of Cancer-Specific Helical and Kinase Domain Mutations of PI3K Alpha. Proc.Natl.Acad.Sci.Usa V. 119 21119 2022.
ISSN: ESSN 1091-6490
PubMed: 36343266
DOI: 10.1073/PNAS.2215621119
Page generated: Wed Jul 16 05:07:12 2025
ISSN: ESSN 1091-6490
PubMed: 36343266
DOI: 10.1073/PNAS.2215621119
Last articles
Fe in 1E1DFe in 1E2R
Fe in 1E29
Fe in 1E0Z
Fe in 1E10
Fe in 1E14
Fe in 1DZ9
Fe in 1DZ6
Fe in 1DZ8
Fe in 1DZ4