Fluorine »
PDB 8gky-8hej »
8gvj »
Fluorine in PDB 8gvj: Crystal Structure of Cmet Kinase Domain Bound By D6808
Enzymatic activity of Crystal Structure of Cmet Kinase Domain Bound By D6808
All present enzymatic activity of Crystal Structure of Cmet Kinase Domain Bound By D6808:
2.7.10.1;
2.7.10.1;
Protein crystallography data
The structure of Crystal Structure of Cmet Kinase Domain Bound By D6808, PDB code: 8gvj
was solved by
Y.H.Chen,
L.Z.Qu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.38 / 2.71 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 66.078, 66.078, 187.302, 90, 90, 90 |
| R / Rfree (%) | 21.9 / 26.2 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of Cmet Kinase Domain Bound By D6808
(pdb code 8gvj). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Crystal Structure of Cmet Kinase Domain Bound By D6808, PDB code: 8gvj:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Crystal Structure of Cmet Kinase Domain Bound By D6808, PDB code: 8gvj:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 8gvj
Go back to
Fluorine binding site 1 out
of 3 in the Crystal Structure of Cmet Kinase Domain Bound By D6808

Mono view
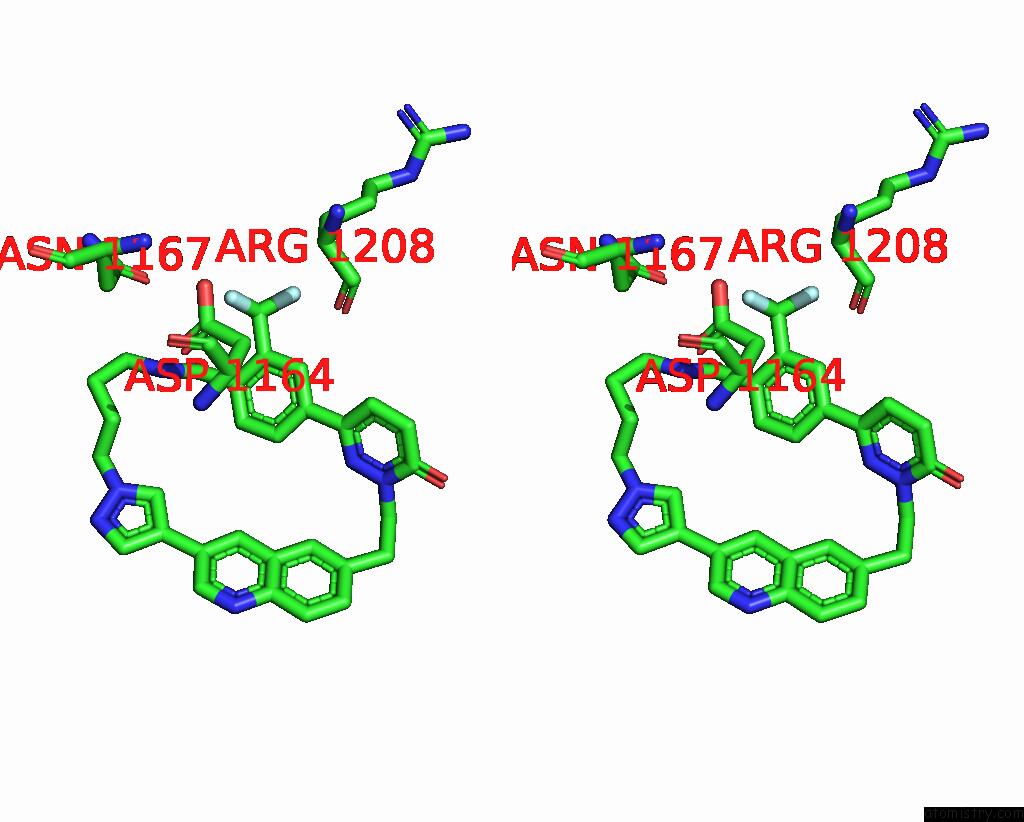
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of Cmet Kinase Domain Bound By D6808 within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 8gvj
Go back to
Fluorine binding site 2 out
of 3 in the Crystal Structure of Cmet Kinase Domain Bound By D6808

Mono view
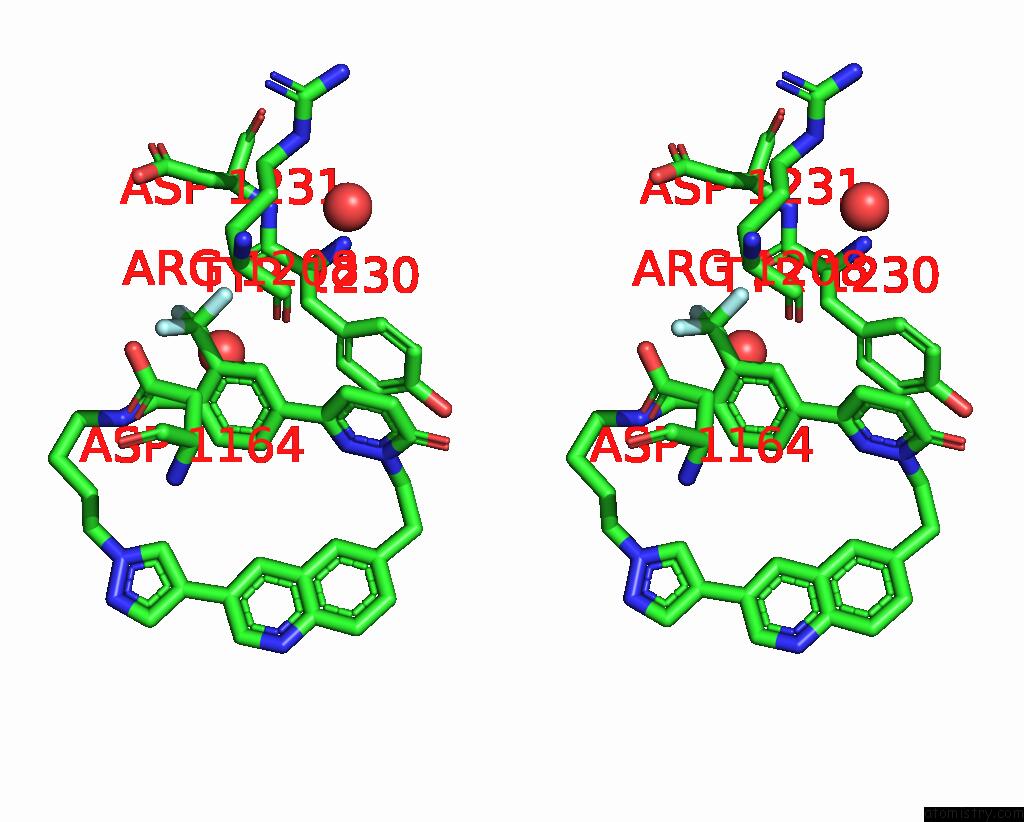
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of Cmet Kinase Domain Bound By D6808 within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 8gvj
Go back to
Fluorine binding site 3 out
of 3 in the Crystal Structure of Cmet Kinase Domain Bound By D6808
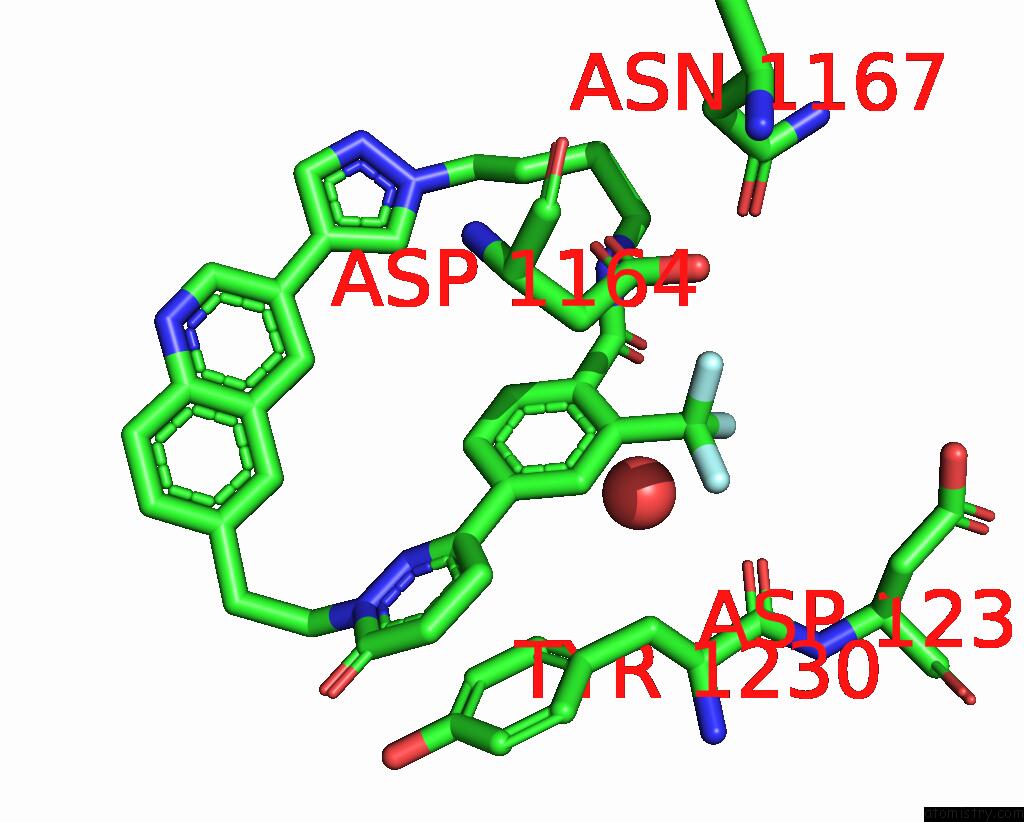
Mono view
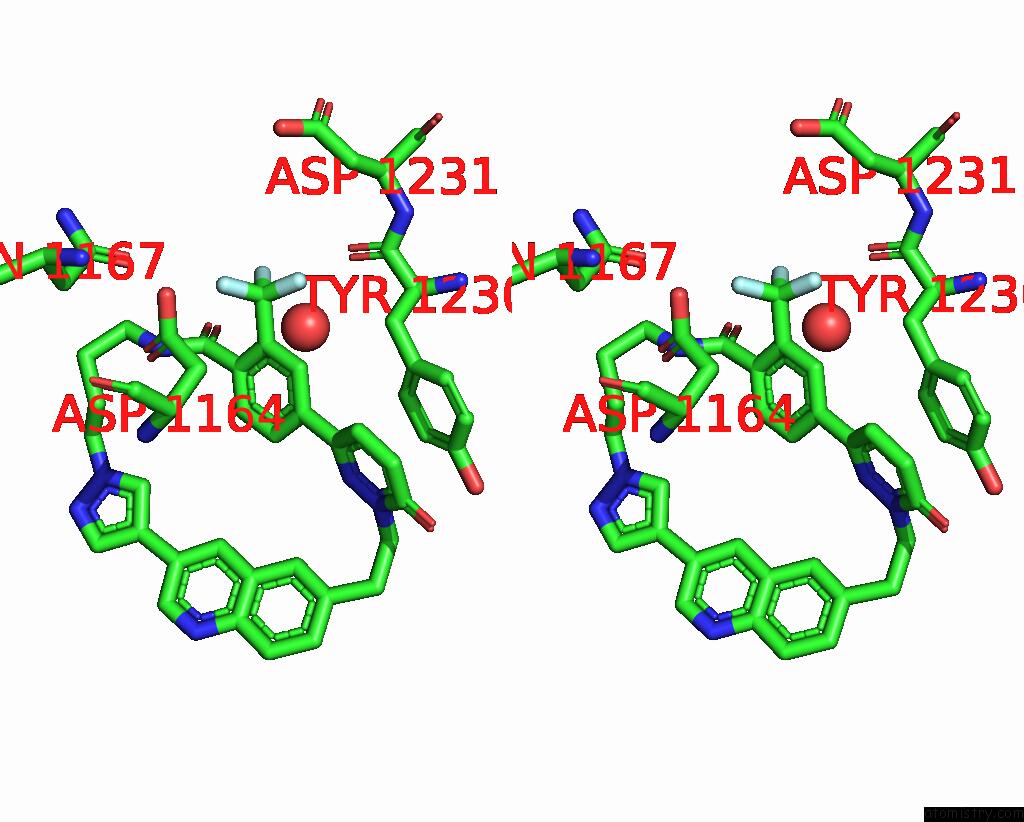
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Crystal Structure of Cmet Kinase Domain Bound By D6808 within 5.0Å range:
|
Reference:
C.Wang,
J.Li,
L.Qu,
X.Tang,
X.Song,
F.Yang,
X.Chen,
Q.Lin,
W.Lin,
Y.Zhou,
Z.Tu,
Y.Chen,
Z.Zhang,
X.Lu.
Discovery of D6808, A Highly Selective and Potent Macrocyclic C-Met Inhibitor For Gastric Cancer Harboring Met Gene Alteration Treatment. J.Med.Chem. V. 65 15140 2022.
ISSN: ISSN 0022-2623
PubMed: 36355693
DOI: 10.1021/ACS.JMEDCHEM.2C00981
Page generated: Wed Jul 16 05:08:28 2025
ISSN: ISSN 0022-2623
PubMed: 36355693
DOI: 10.1021/ACS.JMEDCHEM.2C00981
Last articles
Fe in 1DY7Fe in 1DXR
Fe in 1DXG
Fe in 1DXD
Fe in 1DXC
Fe in 1DWL
Fe in 1DWX
Fe in 1DWW
Fe in 1DWV
Fe in 1DWT