Fluorine »
PDB 9cdl-9f2h »
9ear »
Fluorine in PDB 9ear: CHD1-Nucleosome Complex (Closed State)
Enzymatic activity of CHD1-Nucleosome Complex (Closed State)
All present enzymatic activity of CHD1-Nucleosome Complex (Closed State):
3.6.4.12;
3.6.4.12;
Fluorine Binding Sites:
The binding sites of Fluorine atom in the CHD1-Nucleosome Complex (Closed State)
(pdb code 9ear). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the CHD1-Nucleosome Complex (Closed State), PDB code: 9ear:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the CHD1-Nucleosome Complex (Closed State), PDB code: 9ear:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 9ear
Go back to
Fluorine binding site 1 out
of 3 in the CHD1-Nucleosome Complex (Closed State)
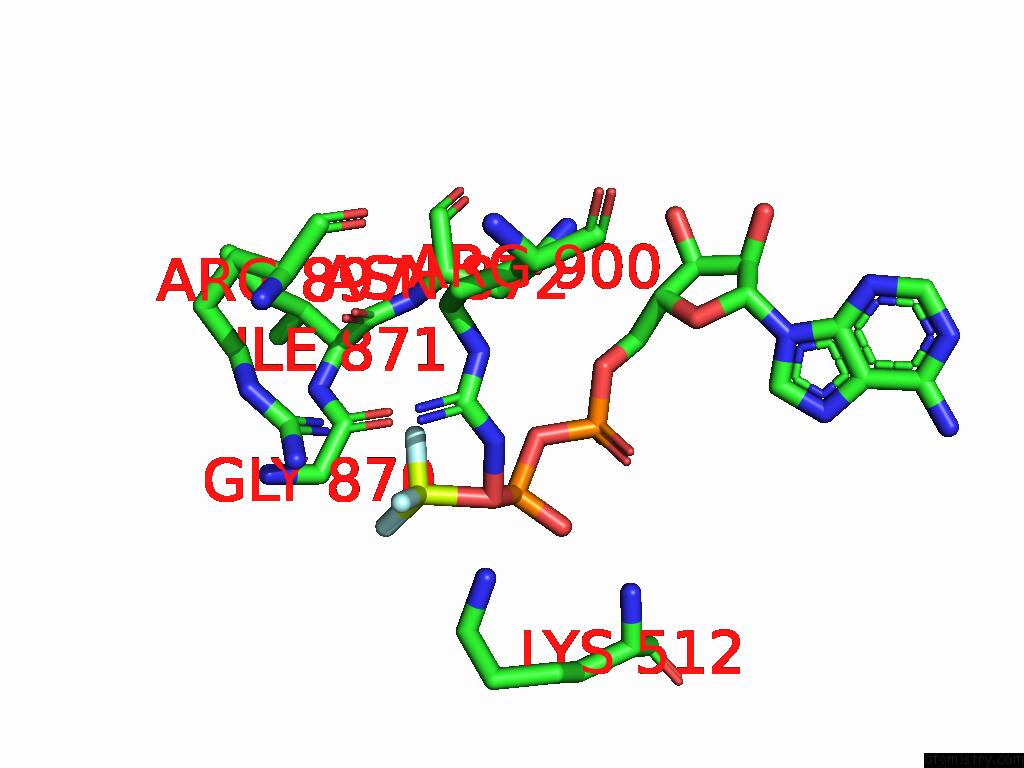
Mono view
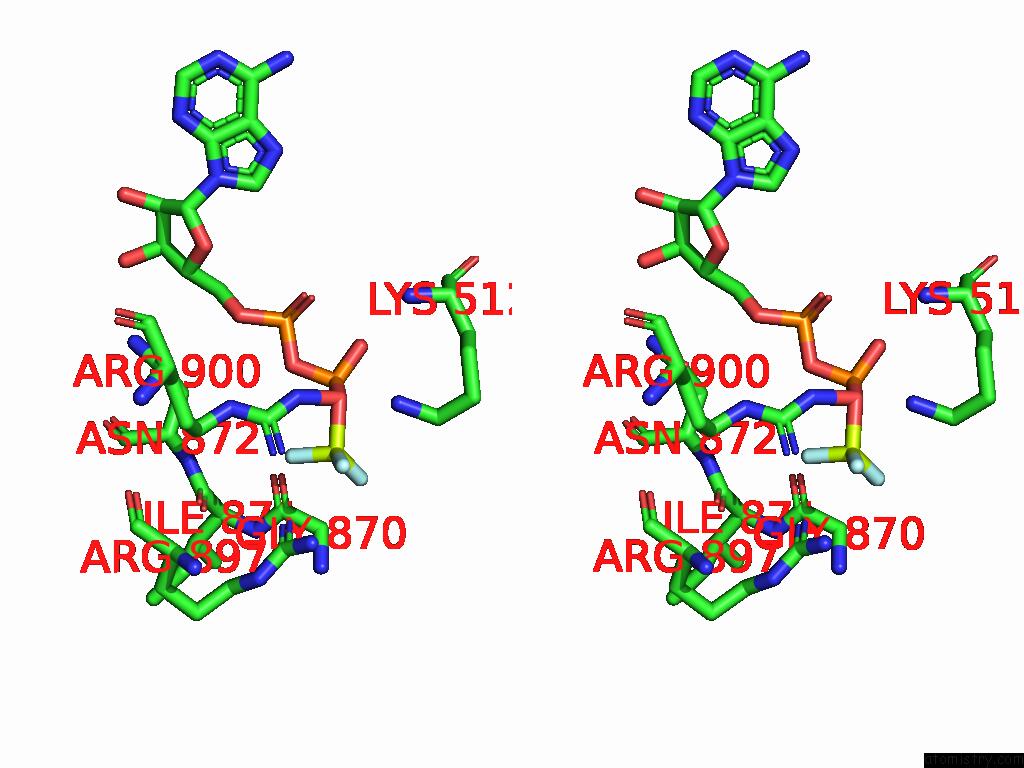
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of CHD1-Nucleosome Complex (Closed State) within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 9ear
Go back to
Fluorine binding site 2 out
of 3 in the CHD1-Nucleosome Complex (Closed State)
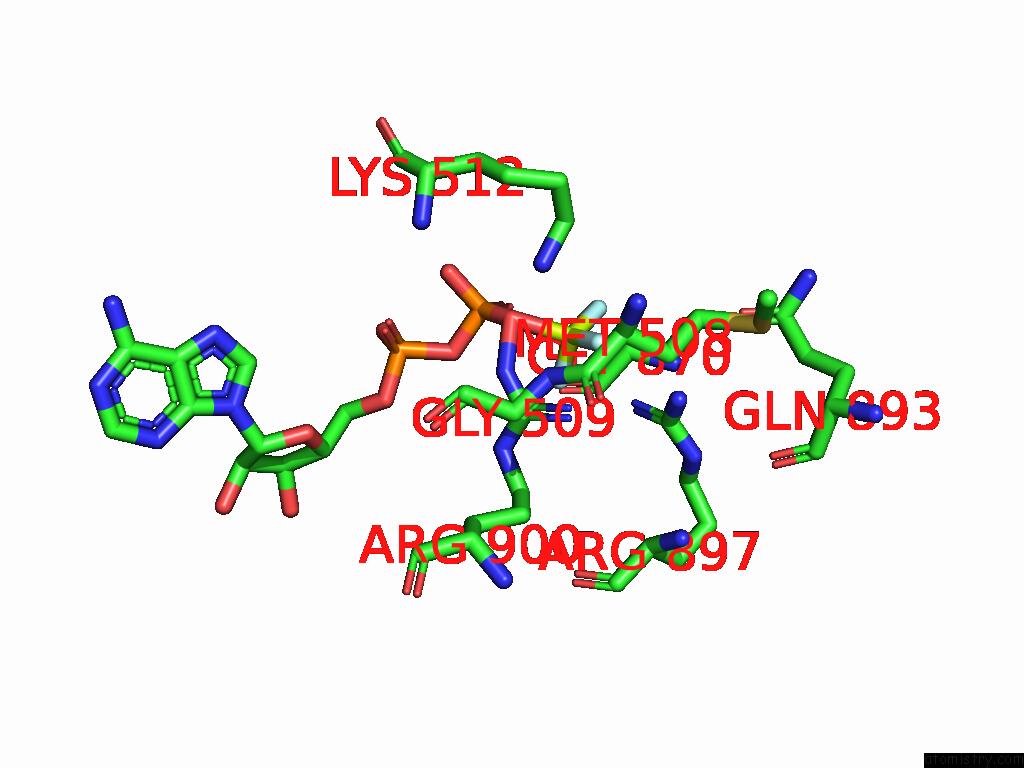
Mono view
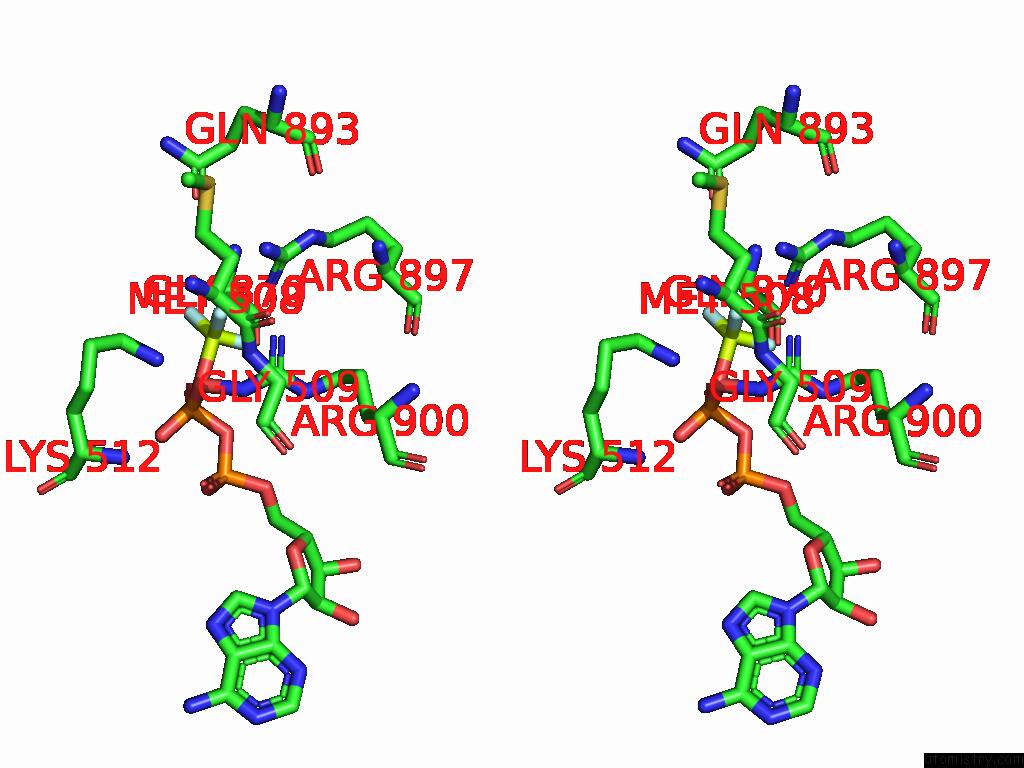
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of CHD1-Nucleosome Complex (Closed State) within 5.0Å range:
|
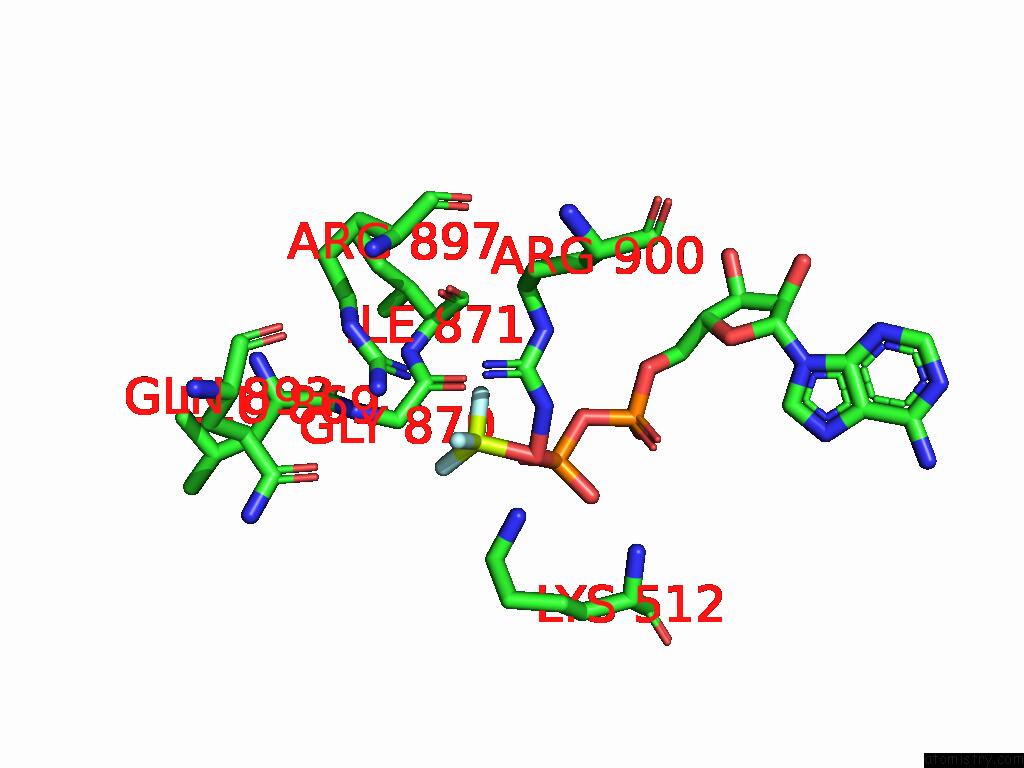
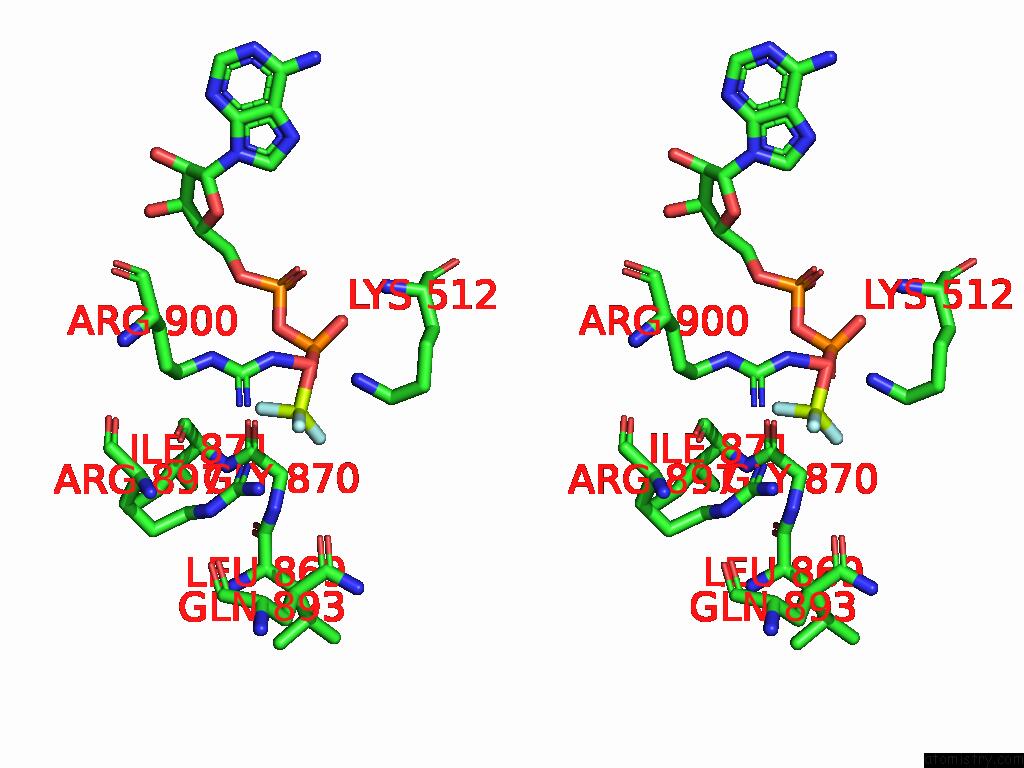
Fluorine binding site 3 out of 3 in 9ear
Go back to
Fluorine binding site 3 out
of 3 in the CHD1-Nucleosome Complex (Closed State)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of CHD1-Nucleosome Complex (Closed State) within 5.0Å range:
|
Reference:
A.M.James,
L.Farnung.
Structural Basis of Human CHD1 Nucleosome Recruitment and Pausing Mol.Cell 2025.
ISSN: ISSN 1097-2765
DOI: 10.1016/J.MOLCEL.2025.04.020
Page generated: Wed Jul 16 10:57:18 2025
ISSN: ISSN 1097-2765
DOI: 10.1016/J.MOLCEL.2025.04.020
Last articles
Fe in 1HGCFe in 1HH5
Fe in 1HHO
Fe in 1HH7
Fe in 1HDS
Fe in 1HGB
Fe in 1HGA
Fe in 1H7W
Fe in 1H7X
Fe in 1HDB