Fluorine »
PDB 9mvy-9u70 »
9mvy »
Fluorine in PDB 9mvy: Crystal Structure of ZMET2 in Complex with Unmethylated Ctg Dna and A Histone H3KC9ME2 Peptide
Enzymatic activity of Crystal Structure of ZMET2 in Complex with Unmethylated Ctg Dna and A Histone H3KC9ME2 Peptide
All present enzymatic activity of Crystal Structure of ZMET2 in Complex with Unmethylated Ctg Dna and A Histone H3KC9ME2 Peptide:
2.1.1.37;
2.1.1.37;
Protein crystallography data
The structure of Crystal Structure of ZMET2 in Complex with Unmethylated Ctg Dna and A Histone H3KC9ME2 Peptide, PDB code: 9mvy
was solved by
G.Herle,
J.Fang,
J.Song,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.40 / 2.71 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 64.515, 277.296, 64.479, 90, 97.26, 90 |
| R / Rfree (%) | 22.7 / 27.1 |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of ZMET2 in Complex with Unmethylated Ctg Dna and A Histone H3KC9ME2 Peptide
(pdb code 9mvy). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Crystal Structure of ZMET2 in Complex with Unmethylated Ctg Dna and A Histone H3KC9ME2 Peptide, PDB code: 9mvy:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Crystal Structure of ZMET2 in Complex with Unmethylated Ctg Dna and A Histone H3KC9ME2 Peptide, PDB code: 9mvy:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 9mvy
Go back to
Fluorine binding site 1 out
of 2 in the Crystal Structure of ZMET2 in Complex with Unmethylated Ctg Dna and A Histone H3KC9ME2 Peptide

Mono view
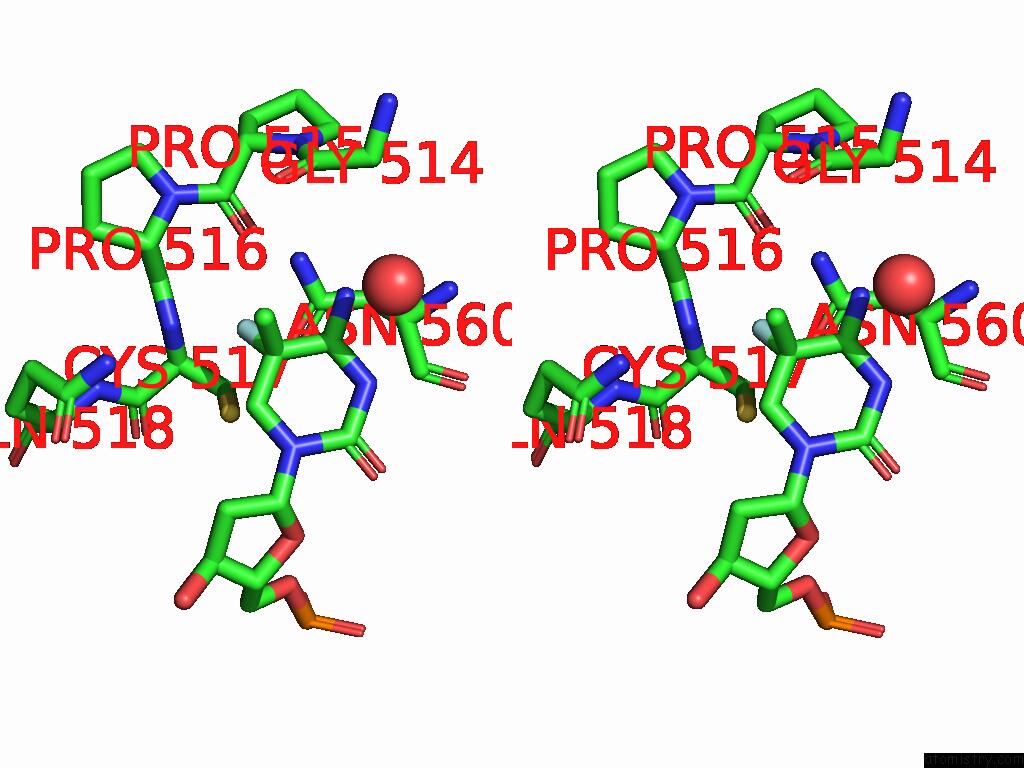
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of ZMET2 in Complex with Unmethylated Ctg Dna and A Histone H3KC9ME2 Peptide within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 9mvy
Go back to
Fluorine binding site 2 out
of 2 in the Crystal Structure of ZMET2 in Complex with Unmethylated Ctg Dna and A Histone H3KC9ME2 Peptide
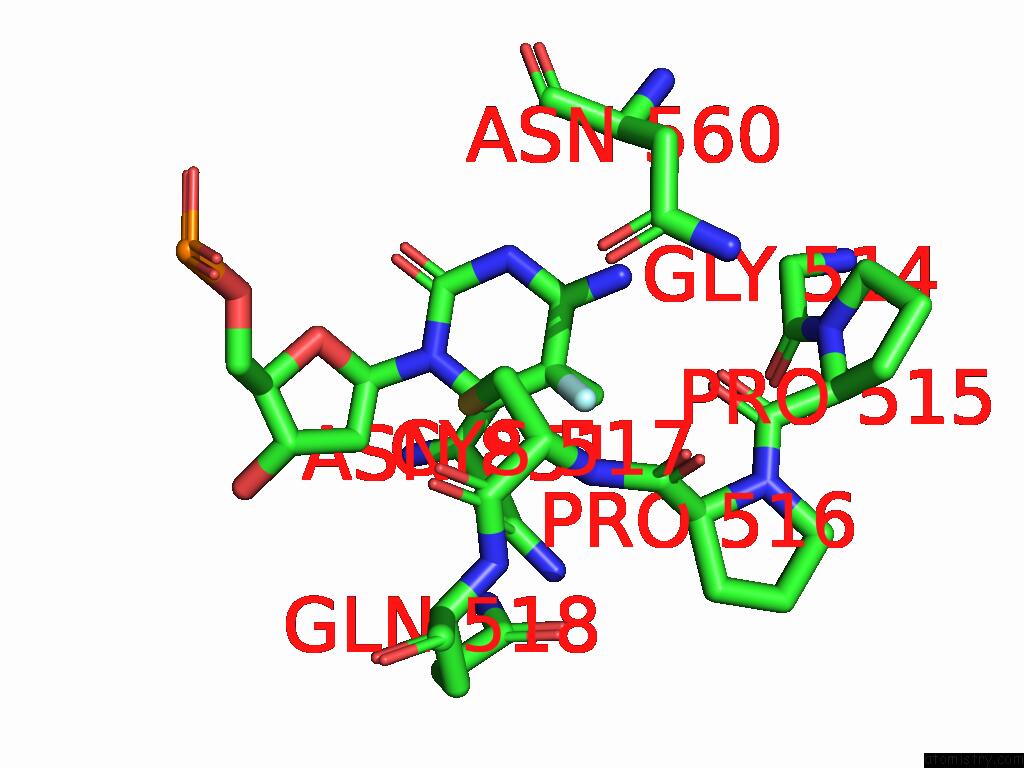
Mono view
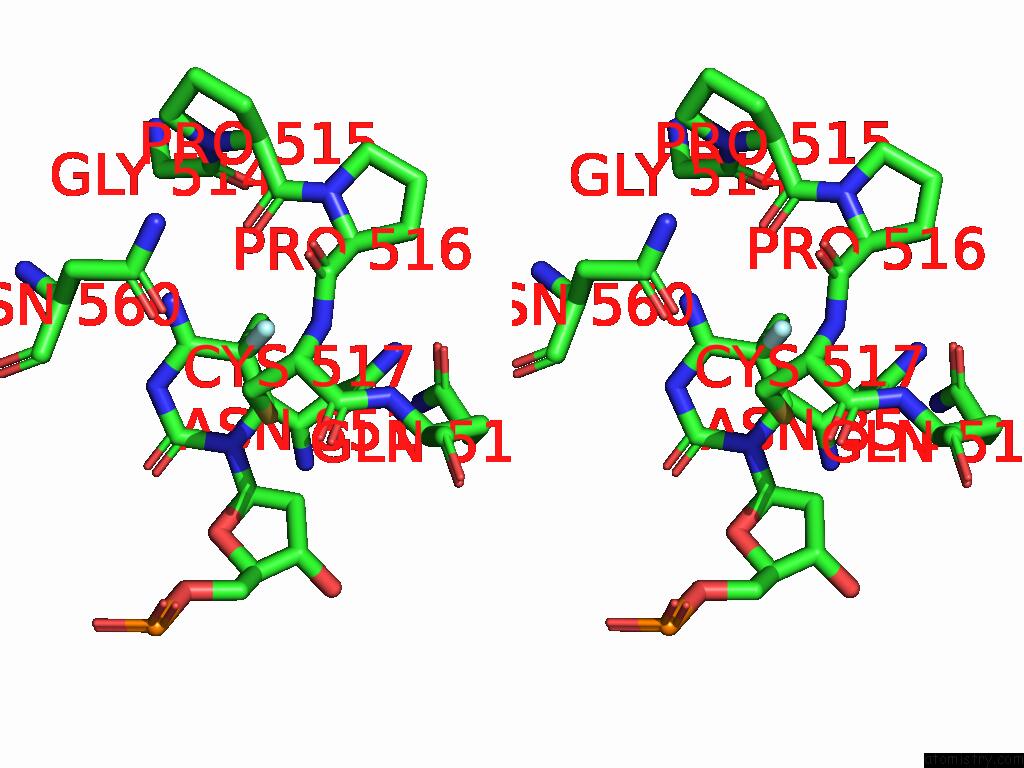
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of ZMET2 in Complex with Unmethylated Ctg Dna and A Histone H3KC9ME2 Peptide within 5.0Å range:
|
Reference:
G.Herle,
J.Fang,
J.Song.
Structure of An Unfavorable De Novo Dna Methylation Complex of Plant Methyltransferase ZMET2. J.Mol.Biol. 69186 2025.
ISSN: ESSN 1089-8638
PubMed: 40335018
DOI: 10.1016/J.JMB.2025.169186
Page generated: Wed Jul 16 11:52:24 2025
ISSN: ESSN 1089-8638
PubMed: 40335018
DOI: 10.1016/J.JMB.2025.169186
Last articles
Fe in 1MJGFe in 1MIO
Fe in 1MKO
Fe in 1MJT
Fe in 1MK8
Fe in 1MDV
Fe in 1MJ4
Fe in 1MHZ
Fe in 1MHY
Fe in 1MG3